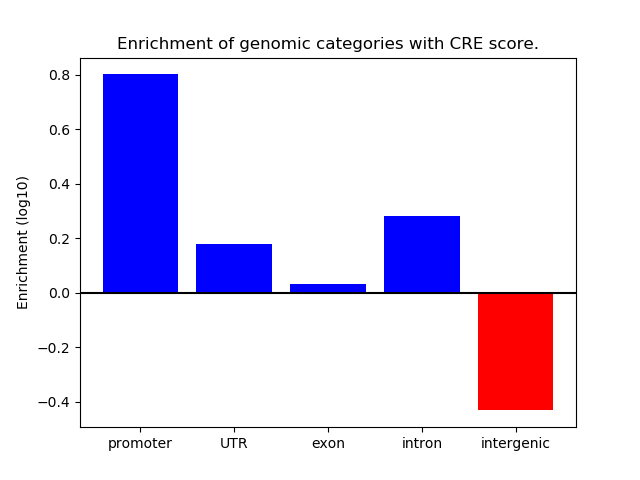
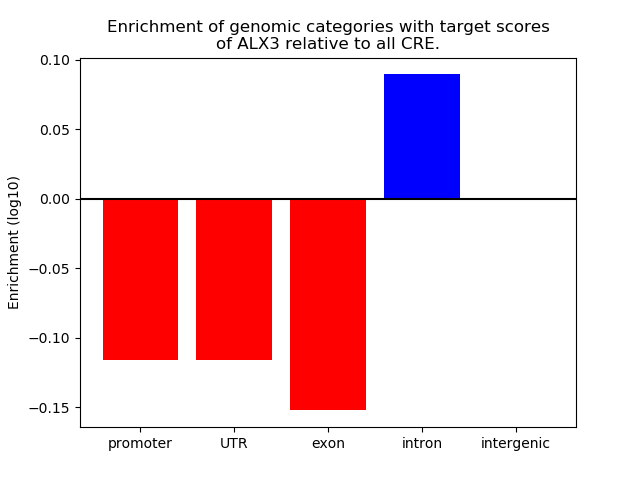
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
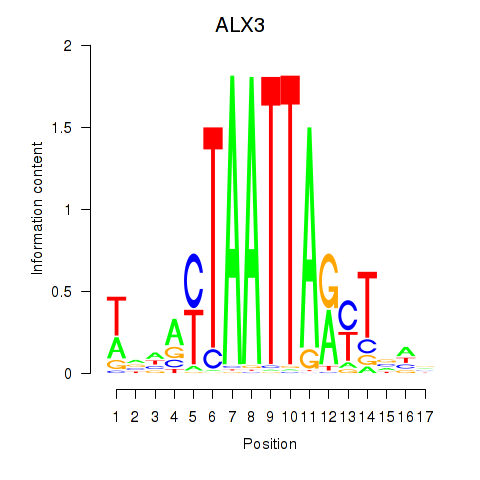
Results for ALX3
Z-value: 0.86
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.6 | ALX homeobox 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_110610926_110611170 | ALX3 | 2274 | 0.202908 | 0.68 | 4.4e-02 | Click! |
| chr1_110612851_110613002 | ALX3 | 396 | 0.780608 | 0.53 | 1.4e-01 | Click! |
| chr1_110610618_110610769 | ALX3 | 2629 | 0.183485 | 0.50 | 1.7e-01 | Click! |
| chr1_110611302_110611453 | ALX3 | 1945 | 0.229462 | 0.44 | 2.3e-01 | Click! |
| chr1_110611525_110611676 | ALX3 | 1722 | 0.254844 | 0.44 | 2.4e-01 | Click! |
Activity of the ALX3 motif across conditions
Conditions sorted by the z-value of the ALX3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_58055562_58056142 | 0.64 |
RP11-479O16.1 |
|
24754 |
0.24 |
| chr7_123390243_123390394 | 0.58 |
RP11-390E23.6 |
|
1196 |
0.34 |
| chr12_66583583_66583798 | 0.52 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
653 |
0.67 |
| chr6_130391698_130391967 | 0.51 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
50130 |
0.15 |
| chr20_21080465_21081932 | 0.49 |
ENSG00000199509 |
. |
38517 |
0.18 |
| chr7_37724366_37724517 | 0.48 |
GPR141 |
G protein-coupled receptor 141 |
966 |
0.42 |
| chr8_95941010_95941161 | 0.42 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
12116 |
0.13 |
| chr7_32929941_32930760 | 0.42 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
415 |
0.87 |
| chrX_78400540_78401325 | 0.39 |
GPR174 |
G protein-coupled receptor 174 |
25537 |
0.27 |
| chr13_41555714_41555943 | 0.39 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
590 |
0.77 |
| chr13_31309966_31310295 | 0.37 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
485 |
0.88 |
| chr1_200865204_200865355 | 0.37 |
C1orf106 |
chromosome 1 open reading frame 106 |
1330 |
0.42 |
| chr11_118134576_118134833 | 0.37 |
MPZL2 |
myelin protein zero-like 2 |
293 |
0.86 |
| chr17_56362337_56362534 | 0.37 |
MPO |
myeloperoxidase |
4139 |
0.15 |
| chr10_30745504_30745655 | 0.37 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
17828 |
0.21 |
| chr21_15917916_15918619 | 0.37 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr5_126094279_126094440 | 0.36 |
ENSG00000252185 |
. |
3250 |
0.29 |
| chr4_147027155_147027306 | 0.36 |
ENSG00000216055 |
. |
16206 |
0.24 |
| chr4_40630076_40630227 | 0.36 |
RBM47 |
RNA binding motif protein 47 |
1730 |
0.47 |
| chr4_36245078_36245884 | 0.36 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
80 |
0.74 |
| chr12_10019991_10020525 | 0.35 |
RP11-290C10.1 |
|
632 |
0.61 |
| chr15_52844361_52844512 | 0.35 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
5505 |
0.2 |
| chr2_103506378_103506656 | 0.35 |
TMEM182 |
transmembrane protein 182 |
128045 |
0.06 |
| chr20_47448799_47448989 | 0.35 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
4474 |
0.31 |
| chr16_88987498_88987922 | 0.35 |
RP11-830F9.7 |
|
16454 |
0.11 |
| chr8_56831708_56831859 | 0.35 |
ENSG00000216204 |
. |
10263 |
0.14 |
| chr2_70780238_70780449 | 0.34 |
TGFA |
transforming growth factor, alpha |
279 |
0.93 |
| chr10_129861850_129862130 | 0.34 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
16156 |
0.26 |
| chr1_8441651_8441802 | 0.34 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
16828 |
0.17 |
| chr6_133069776_133069927 | 0.34 |
RP1-55C23.7 |
|
3963 |
0.15 |
| chr16_31271922_31272073 | 0.34 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
682 |
0.53 |
| chr22_40858929_40859404 | 0.34 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
256 |
0.92 |
| chr20_52271481_52272252 | 0.33 |
ENSG00000238468 |
. |
13431 |
0.22 |
| chr2_61111507_61111691 | 0.33 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
2808 |
0.27 |
| chr13_73631347_73631498 | 0.33 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
1508 |
0.47 |
| chr1_234735009_234735261 | 0.32 |
ENSG00000212144 |
. |
6114 |
0.19 |
| chr12_10281805_10282201 | 0.32 |
CLEC7A |
C-type lectin domain family 7, member A |
678 |
0.6 |
| chr12_27150397_27150548 | 0.32 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
2271 |
0.24 |
| chr3_187491387_187491633 | 0.32 |
BCL6 |
B-cell CLL/lymphoma 6 |
27995 |
0.2 |
| chr5_68905149_68905341 | 0.32 |
RP11-848G14.2 |
|
8895 |
0.19 |
| chr9_93557702_93558119 | 0.32 |
SYK |
spleen tyrosine kinase |
6159 |
0.34 |
| chr5_67562966_67563283 | 0.32 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
12944 |
0.27 |
| chr15_52023462_52024365 | 0.31 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
6405 |
0.15 |
| chr5_57787350_57787738 | 0.31 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
280 |
0.92 |
| chr15_69074948_69075145 | 0.31 |
ENSG00000265195 |
. |
19218 |
0.22 |
| chr4_105887659_105887892 | 0.31 |
ENSG00000251906 |
. |
8044 |
0.31 |
| chr11_128589269_128589524 | 0.31 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chr14_75885513_75886422 | 0.31 |
RP11-293M10.6 |
|
8426 |
0.2 |
| chr2_153267501_153267713 | 0.30 |
FMNL2 |
formin-like 2 |
75856 |
0.12 |
| chr7_37726318_37726469 | 0.30 |
GPR141 |
G protein-coupled receptor 141 |
2918 |
0.28 |
| chr21_39630264_39630415 | 0.30 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
1469 |
0.53 |
| chr1_158802859_158803010 | 0.30 |
MNDA |
myeloid cell nuclear differentiation antigen |
1827 |
0.35 |
| chr5_70314152_70314347 | 0.30 |
NAIP |
NLR family, apoptosis inhibitory protein |
2288 |
0.36 |
| chr15_43531850_43532099 | 0.30 |
ENSG00000202211 |
. |
7904 |
0.13 |
| chr5_17282868_17283019 | 0.30 |
ENSG00000252908 |
. |
42223 |
0.15 |
| chr2_64864648_64864996 | 0.29 |
SERTAD2 |
SERTA domain containing 2 |
16225 |
0.22 |
| chr2_70157637_70157788 | 0.29 |
MXD1 |
MAX dimerization protein 1 |
15392 |
0.11 |
| chr12_8666250_8666401 | 0.29 |
CLEC4D |
C-type lectin domain family 4, member D |
189 |
0.93 |
| chr10_12064652_12064803 | 0.29 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
13169 |
0.19 |
| chr11_33284734_33284885 | 0.29 |
HIPK3 |
homeodomain interacting protein kinase 3 |
4932 |
0.25 |
| chr5_69760327_69760484 | 0.29 |
RP11-497H16.7 |
|
8870 |
0.28 |
| chr7_115668295_115668446 | 0.29 |
TFEC |
transcription factor EC |
2425 |
0.43 |
| chr7_105912156_105912307 | 0.29 |
NAMPT |
nicotinamide phosphoribosyltransferase |
13136 |
0.21 |
| chr9_77757026_77757296 | 0.29 |
ENSG00000200041 |
. |
39376 |
0.14 |
| chr11_640631_640896 | 0.29 |
DRD4 |
dopamine receptor D4 |
3470 |
0.1 |
| chr2_202076992_202077403 | 0.29 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
20969 |
0.15 |
| chr2_158299843_158300108 | 0.29 |
CYTIP |
cytohesin 1 interacting protein |
679 |
0.68 |
| chr3_98480702_98481101 | 0.29 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
1260 |
0.48 |
| chr1_112013416_112013602 | 0.28 |
C1orf162 |
chromosome 1 open reading frame 162 |
2905 |
0.14 |
| chr7_106507057_106507208 | 0.28 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1208 |
0.59 |
| chr1_36500237_36500507 | 0.28 |
RP4-665N4.8 |
|
21141 |
0.16 |
| chrX_133523066_133523217 | 0.28 |
ENSG00000251962 |
. |
14271 |
0.17 |
| chr8_27223967_27224118 | 0.27 |
PTK2B |
protein tyrosine kinase 2 beta |
14126 |
0.22 |
| chr19_2267105_2267256 | 0.27 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
2305 |
0.12 |
| chr13_68202936_68203087 | 0.27 |
PCDH9 |
protocadherin 9 |
398543 |
0.01 |
| chr4_1581349_1582263 | 0.27 |
FAM53A |
family with sequence similarity 53, member A |
75329 |
0.08 |
| chr12_64852912_64853865 | 0.27 |
TBK1 |
TANK-binding kinase 1 |
7133 |
0.2 |
| chr3_134261756_134261907 | 0.27 |
CEP63 |
centrosomal protein 63kDa |
3167 |
0.29 |
| chr20_48328141_48328292 | 0.27 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
2199 |
0.3 |
| chr11_46581548_46581763 | 0.27 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
31259 |
0.15 |
| chr8_112438072_112438223 | 0.26 |
ENSG00000222146 |
. |
722907 |
0.0 |
| chr13_32616629_32616780 | 0.26 |
FRY-AS1 |
FRY antisense RNA 1 |
10928 |
0.22 |
| chr7_87503109_87503295 | 0.26 |
DBF4 |
DBF4 homolog (S. cerevisiae) |
2342 |
0.29 |
| chr3_47123794_47124082 | 0.26 |
ENSG00000251938 |
. |
6160 |
0.22 |
| chr1_207228136_207228287 | 0.26 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
166 |
0.92 |
| chr10_17269607_17270502 | 0.26 |
VIM |
vimentin |
204 |
0.89 |
| chr17_29636179_29636457 | 0.26 |
EVI2B |
ecotropic viral integration site 2B |
4784 |
0.15 |
| chr4_84161976_84162127 | 0.26 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
43867 |
0.16 |
| chr3_187492063_187492306 | 0.25 |
BCL6 |
B-cell CLL/lymphoma 6 |
28669 |
0.2 |
| chr9_129253296_129253447 | 0.25 |
ENSG00000221768 |
. |
39899 |
0.15 |
| chr16_48411039_48411308 | 0.25 |
SIAH1 |
siah E3 ubiquitin protein ligase 1 |
8188 |
0.18 |
| chr17_78755045_78755196 | 0.25 |
RP11-28G8.1 |
|
24312 |
0.21 |
| chr12_64798382_64799443 | 0.25 |
XPOT |
exportin, tRNA |
86 |
0.97 |
| chr8_22961002_22961301 | 0.25 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
717 |
0.58 |
| chr2_103048870_103049021 | 0.25 |
ENSG00000264764 |
. |
196 |
0.9 |
| chr21_36772703_36772854 | 0.25 |
ENSG00000211590 |
. |
320235 |
0.01 |
| chr7_105953972_105954204 | 0.25 |
NAMPT |
nicotinamide phosphoribosyltransferase |
27316 |
0.2 |
| chr2_207998546_207999225 | 0.25 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
24 |
0.98 |
| chr22_31615175_31615326 | 0.24 |
ENSG00000199695 |
. |
3589 |
0.13 |
| chr6_11350314_11350465 | 0.24 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
32143 |
0.22 |
| chr13_86159310_86159559 | 0.24 |
ENSG00000207012 |
. |
446 |
0.91 |
| chr22_17598033_17598230 | 0.24 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
4012 |
0.17 |
| chr3_138524893_138525044 | 0.24 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
28626 |
0.22 |
| chr12_32742094_32742245 | 0.24 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
9291 |
0.26 |
| chr5_76112526_76112677 | 0.24 |
F2RL1 |
coagulation factor II (thrombin) receptor-like 1 |
2157 |
0.27 |
| chr1_192776419_192777001 | 0.24 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
1461 |
0.52 |
| chr2_182168845_182168996 | 0.24 |
ENSG00000266705 |
. |
1459 |
0.58 |
| chrX_19703578_19703729 | 0.24 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
14537 |
0.29 |
| chr11_33912422_33912614 | 0.24 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
1318 |
0.47 |
| chr10_70850849_70851349 | 0.24 |
SRGN |
serglycin |
3225 |
0.25 |
| chr20_52370901_52371111 | 0.24 |
ENSG00000238468 |
. |
85709 |
0.09 |
| chr7_17039488_17039788 | 0.24 |
AGR3 |
anterior gradient 3 |
118027 |
0.06 |
| chr7_20825937_20826584 | 0.24 |
SP8 |
Sp8 transcription factor |
245 |
0.96 |
| chr18_3453699_3454909 | 0.24 |
TGIF1 |
TGFB-induced factor homeobox 1 |
532 |
0.8 |
| chr8_26184784_26184935 | 0.24 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
33860 |
0.18 |
| chr8_56792958_56793527 | 0.24 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
848 |
0.55 |
| chr11_72700714_72700865 | 0.23 |
FCHSD2 |
FCH and double SH3 domains 2 |
732 |
0.75 |
| chr12_25208788_25208939 | 0.23 |
LRMP |
lymphoid-restricted membrane protein |
3189 |
0.26 |
| chr15_38376930_38377081 | 0.23 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
149547 |
0.04 |
| chr1_226251298_226251924 | 0.23 |
H3F3A |
H3 histone, family 3A |
67 |
0.97 |
| chr10_17395409_17395560 | 0.23 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
26376 |
0.16 |
| chr1_221173678_221173829 | 0.23 |
HLX |
H2.0-like homeobox |
119169 |
0.06 |
| chr7_133456734_133456885 | 0.23 |
EXOC4 |
exocyst complex component 4 |
158413 |
0.04 |
| chr6_159071306_159071615 | 0.23 |
SYTL3 |
synaptotagmin-like 3 |
414 |
0.83 |
| chr9_132807889_132808048 | 0.23 |
FNBP1 |
formin binding protein 1 |
2495 |
0.28 |
| chr21_30543274_30543425 | 0.23 |
ENSG00000212479 |
. |
9611 |
0.13 |
| chr8_131206424_131206650 | 0.23 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
13429 |
0.23 |
| chr20_1550627_1550778 | 0.23 |
SIRPB1 |
signal-regulatory protein beta 1 |
1856 |
0.26 |
| chr1_42230520_42230906 | 0.23 |
ENSG00000264896 |
. |
5901 |
0.3 |
| chr14_70111468_70111619 | 0.22 |
KIAA0247 |
KIAA0247 |
33230 |
0.18 |
| chr2_178249081_178249232 | 0.22 |
AGPS |
alkylglycerone phosphate synthase |
8216 |
0.14 |
| chr2_201238626_201239302 | 0.22 |
ENSG00000201649 |
. |
1066 |
0.5 |
| chr6_18391062_18391213 | 0.22 |
RNF144B |
ring finger protein 144B |
3556 |
0.26 |
| chr16_50059050_50060305 | 0.22 |
CNEP1R1 |
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
246 |
0.93 |
| chr21_36367168_36367319 | 0.22 |
RUNX1 |
runt-related transcription factor 1 |
54219 |
0.18 |
| chr7_5594421_5594572 | 0.22 |
CTB-161C1.1 |
|
1866 |
0.25 |
| chr6_107297762_107297913 | 0.22 |
C6orf203 |
chromosome 6 open reading frame 203 |
51570 |
0.14 |
| chr19_6425167_6425318 | 0.22 |
KHSRP |
KH-type splicing regulatory protein |
437 |
0.66 |
| chr9_139434770_139435052 | 0.22 |
RP11-413M3.4 |
|
2422 |
0.15 |
| chr7_37011253_37011404 | 0.22 |
ELMO1 |
engulfment and cell motility 1 |
13337 |
0.2 |
| chr6_41528112_41529137 | 0.22 |
FOXP4 |
forkhead box P4 |
4863 |
0.19 |
| chr8_12589361_12589512 | 0.22 |
LONRF1 |
LON peptidase N-terminal domain and ring finger 1 |
2540 |
0.26 |
| chr10_119644806_119644957 | 0.22 |
RP11-354M20.3 |
|
132117 |
0.05 |
| chr2_215664874_215665025 | 0.22 |
BARD1 |
BRCA1 associated RING domain 1 |
9448 |
0.21 |
| chr6_134551187_134551338 | 0.22 |
ENSG00000238631 |
. |
23610 |
0.16 |
| chr5_148189087_148189390 | 0.22 |
ADRB2 |
adrenoceptor beta 2, surface |
16918 |
0.25 |
| chr10_70087727_70088191 | 0.21 |
HNRNPH3 |
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
3873 |
0.2 |
| chr20_46412966_46413363 | 0.21 |
SULF2 |
sulfatase 2 |
1069 |
0.59 |
| chr7_50348449_50348781 | 0.21 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
297 |
0.94 |
| chr10_126664611_126664762 | 0.21 |
RP11-298J20.3 |
|
29685 |
0.14 |
| chr13_71409756_71409907 | 0.21 |
ENSG00000202433 |
. |
376235 |
0.01 |
| chr3_50658137_50658523 | 0.21 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
3509 |
0.19 |
| chr3_108988766_108988917 | 0.21 |
ENSG00000252889 |
. |
44865 |
0.15 |
| chr1_200987607_200987758 | 0.21 |
KIF21B |
kinesin family member 21B |
4854 |
0.21 |
| chr15_45940670_45940821 | 0.21 |
SQRDL |
sulfide quinone reductase-like (yeast) |
2666 |
0.28 |
| chr13_78434617_78434768 | 0.21 |
EDNRB |
endothelin receptor type B |
58253 |
0.12 |
| chr12_54891959_54892179 | 0.21 |
NCKAP1L |
NCK-associated protein 1-like |
499 |
0.73 |
| chr1_173807426_173807577 | 0.21 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
13704 |
0.1 |
| chr8_9005081_9005340 | 0.21 |
PPP1R3B |
protein phosphatase 1, regulatory subunit 3B |
2996 |
0.21 |
| chr15_94796301_94796468 | 0.21 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
21617 |
0.29 |
| chr7_139555611_139555822 | 0.21 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
26606 |
0.23 |
| chr3_122029046_122029197 | 0.21 |
CSTA |
cystatin A (stefin A) |
14970 |
0.16 |
| chr11_59949160_59949311 | 0.21 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
96 |
0.97 |
| chr1_169679248_169679951 | 0.21 |
SELL |
selectin L |
1240 |
0.48 |
| chr12_12510418_12511218 | 0.21 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
466 |
0.63 |
| chr5_39412000_39412151 | 0.21 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
12895 |
0.26 |
| chr22_36843986_36844137 | 0.21 |
ENSG00000252225 |
. |
7046 |
0.15 |
| chr3_194821900_194822051 | 0.21 |
XXYLT1-AS1 |
XXYLT1 antisense RNA 1 |
6658 |
0.17 |
| chr4_139635357_139635508 | 0.21 |
ENSG00000238971 |
. |
97488 |
0.08 |
| chr1_225610206_225610357 | 0.21 |
LBR |
lamin B receptor |
5318 |
0.2 |
| chr11_14375529_14375805 | 0.21 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
379 |
0.91 |
| chr3_58341767_58341918 | 0.21 |
PXK |
PX domain containing serine/threonine kinase |
22809 |
0.17 |
| chr9_85107233_85107384 | 0.21 |
SPATA31D1 |
SPATA31 subfamily D, member 1 |
503621 |
0.0 |
| chr19_13208223_13208471 | 0.20 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
5334 |
0.11 |
| chr4_68636095_68636246 | 0.20 |
GNRHR |
gonadotropin-releasing hormone receptor |
16092 |
0.17 |
| chr5_96297085_96297358 | 0.20 |
ENSG00000200884 |
. |
1240 |
0.43 |
| chr13_99910339_99910626 | 0.20 |
GPR18 |
G protein-coupled receptor 18 |
146 |
0.96 |
| chr19_19083033_19083500 | 0.20 |
ENSG00000241172 |
. |
14199 |
0.1 |
| chr14_88471433_88472132 | 0.20 |
GPR65 |
G protein-coupled receptor 65 |
314 |
0.88 |
| chr9_141104243_141104394 | 0.20 |
TUBBP5 |
tubulin, beta pseudogene 5 |
34821 |
0.19 |
| chr8_131290835_131290986 | 0.20 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
31622 |
0.23 |
| chr7_83932213_83932364 | 0.20 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
107527 |
0.08 |
| chr1_29253817_29254129 | 0.20 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12882 |
0.18 |
| chr2_148386736_148386887 | 0.20 |
ENSG00000253083 |
. |
84312 |
0.09 |
| chr4_87930908_87931059 | 0.20 |
AFF1 |
AF4/FMR2 family, member 1 |
2562 |
0.39 |
| chr9_20490881_20491032 | 0.20 |
ENSG00000264941 |
. |
11384 |
0.23 |
| chr4_79473579_79473973 | 0.20 |
ANXA3 |
annexin A3 |
655 |
0.79 |
| chr17_56736328_56736479 | 0.20 |
ENSG00000202077 |
. |
263 |
0.86 |
| chr15_99444137_99444307 | 0.20 |
RP11-654A16.1 |
|
7458 |
0.22 |
| chr8_102506754_102506905 | 0.20 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
1843 |
0.34 |
| chr9_101881133_101881552 | 0.20 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
8715 |
0.21 |
| chr2_149863937_149864088 | 0.20 |
LYPD6B |
LY6/PLAUR domain containing 6B |
30969 |
0.22 |
| chr6_6396318_6396469 | 0.20 |
F13A1 |
coagulation factor XIII, A1 polypeptide |
75147 |
0.11 |
| chr12_100522777_100522928 | 0.20 |
UHRF1BP1L |
UHRF1 binding protein 1-like |
13773 |
0.13 |
| chr3_15336001_15336152 | 0.20 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
18524 |
0.13 |
| chr3_136557614_136557868 | 0.20 |
RP11-731C17.2 |
|
122 |
0.96 |
| chr5_88484775_88484926 | 0.20 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
223158 |
0.02 |
| chr14_68320261_68320412 | 0.20 |
RAD51B |
RAD51 paralog B |
30075 |
0.16 |
| chr2_70368629_70369219 | 0.20 |
C2orf42 |
chromosome 2 open reading frame 42 |
40203 |
0.11 |
| chr14_32555117_32555268 | 0.20 |
ARHGAP5 |
Rho GTPase activating protein 5 |
7755 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.2 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.2 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.1 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 0.2 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.2 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.2 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0002902 | regulation of B cell apoptotic process(GO:0002902) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0071675 | mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0002669 | lymphocyte anergy(GO:0002249) T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.0 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0044116 | growth involved in symbiotic interaction(GO:0044110) growth of symbiont involved in interaction with host(GO:0044116) growth of symbiont in host(GO:0044117) regulation of growth of symbiont in host(GO:0044126) negative regulation of growth of symbiont in host(GO:0044130) modulation of growth of symbiont involved in interaction with host(GO:0044144) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.0 | GO:0055022 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.3 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.1 | GO:0048668 | collateral sprouting(GO:0048668) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.4 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.2 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.1 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |