Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
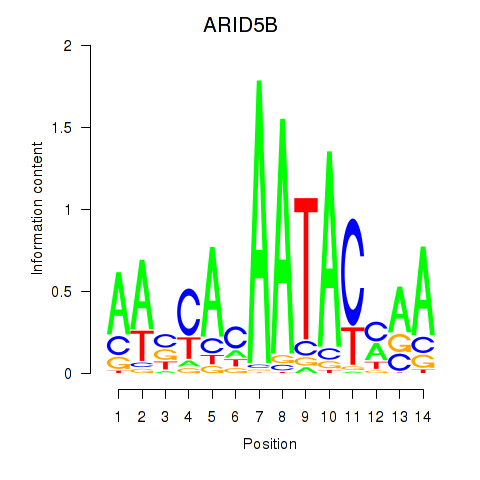
Results for ARID5B
Z-value: 1.60
Transcription factors associated with ARID5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5B
|
ENSG00000150347.10 | AT-rich interaction domain 5B |
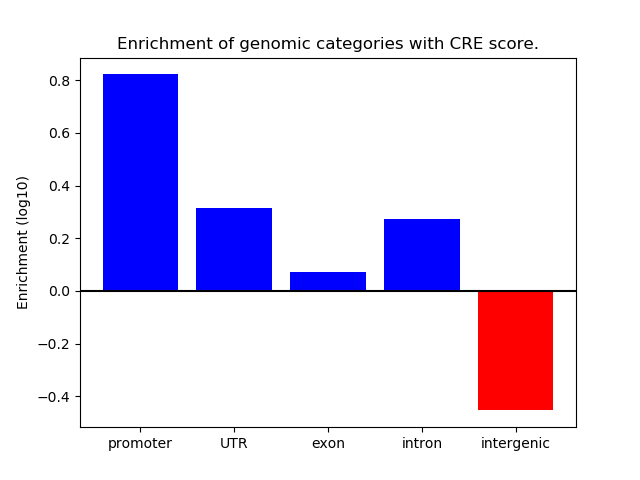
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_63598059_63598210 | ARID5B | 62925 | 0.132995 | 0.77 | 1.4e-02 | Click! |
| chr10_63816154_63816305 | ARID5B | 7259 | 0.280119 | 0.75 | 1.9e-02 | Click! |
| chr10_63785952_63786103 | ARID5B | 22943 | 0.242276 | 0.75 | 2.1e-02 | Click! |
| chr10_63656807_63656958 | ARID5B | 4177 | 0.295667 | -0.68 | 4.3e-02 | Click! |
| chr10_63597664_63597815 | ARID5B | 63320 | 0.132152 | 0.67 | 4.7e-02 | Click! |
Activity of the ARID5B motif across conditions
Conditions sorted by the z-value of the ARID5B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
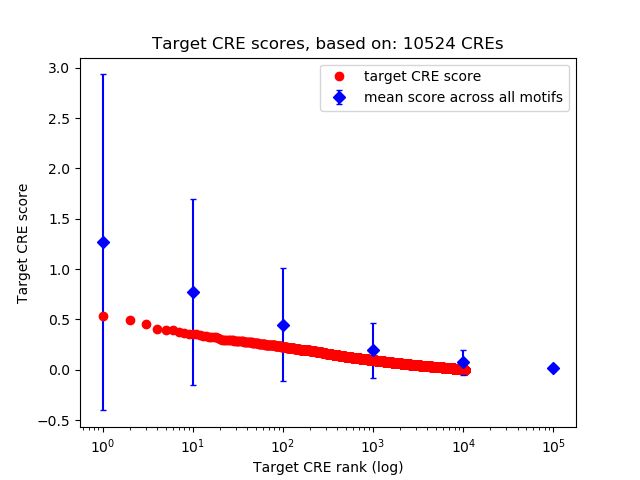
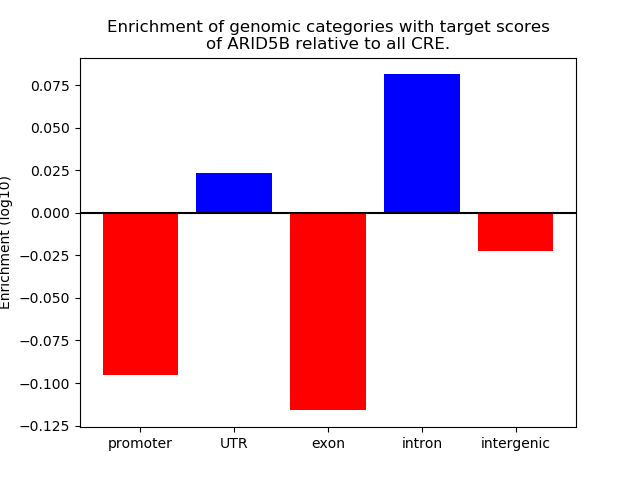
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr21_36256728_36257172 | 0.54 |
RUNX1 |
runt-related transcription factor 1 |
2530 |
0.42 |
| chr21_35133624_35133954 | 0.50 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
26443 |
0.2 |
| chr8_126013201_126013598 | 0.45 |
SQLE |
squalene epoxidase |
2613 |
0.23 |
| chr4_177874509_177874660 | 0.40 |
VEGFC |
vascular endothelial growth factor C |
160703 |
0.04 |
| chr8_116594269_116594420 | 0.39 |
TRPS1 |
trichorhinophalangeal syndrome I |
37953 |
0.21 |
| chr8_102802440_102802591 | 0.39 |
NCALD |
neurocalcin delta |
660 |
0.79 |
| chr4_57975072_57975270 | 0.37 |
IGFBP7-AS1 |
IGFBP7 antisense RNA 1 |
757 |
0.52 |
| chr13_96704851_96705002 | 0.36 |
UGGT2 |
UDP-glucose glycoprotein glucosyltransferase 2 |
711 |
0.77 |
| chr8_81400855_81401257 | 0.36 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
2608 |
0.27 |
| chr13_33162802_33163103 | 0.36 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
2388 |
0.38 |
| chr4_16899723_16900066 | 0.36 |
LDB2 |
LIM domain binding 2 |
291 |
0.95 |
| chr18_18944170_18944321 | 0.35 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
691 |
0.74 |
| chr17_66951279_66951576 | 0.34 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
45 |
0.98 |
| chr14_55239224_55239463 | 0.34 |
SAMD4A |
sterile alpha motif domain containing 4A |
17792 |
0.24 |
| chr2_190042021_190042178 | 0.33 |
COL5A2 |
collagen, type V, alpha 2 |
2506 |
0.34 |
| chr3_16219029_16219297 | 0.33 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
2947 |
0.33 |
| chr21_17652326_17652785 | 0.32 |
ENSG00000201025 |
. |
4534 |
0.36 |
| chr14_101292536_101293492 | 0.32 |
AL117190.2 |
|
2523 |
0.08 |
| chr12_115105992_115106327 | 0.32 |
ENSG00000199220 |
. |
10292 |
0.22 |
| chr8_12986829_12986980 | 0.31 |
DLC1 |
deleted in liver cancer 1 |
4079 |
0.28 |
| chrX_101054612_101054832 | 0.30 |
NXF5 |
nuclear RNA export factor 5 |
43449 |
0.14 |
| chr4_89978098_89978439 | 0.30 |
FAM13A |
family with sequence similarity 13, member A |
43 |
0.98 |
| chr4_77610213_77610516 | 0.30 |
AC107072.2 |
|
51588 |
0.12 |
| chr5_99930860_99931067 | 0.30 |
FAM174A |
family with sequence similarity 174, member A |
59954 |
0.16 |
| chr10_264410_264561 | 0.30 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
8594 |
0.22 |
| chr6_46292394_46292545 | 0.29 |
RCAN2 |
regulator of calcineurin 2 |
936 |
0.7 |
| chr1_156540904_156541055 | 0.29 |
IQGAP3 |
IQ motif containing GTPase activating protein 3 |
1353 |
0.26 |
| chr5_98111208_98111383 | 0.29 |
RGMB |
repulsive guidance molecule family member b |
1956 |
0.33 |
| chr1_103571993_103572213 | 0.29 |
COL11A1 |
collagen, type XI, alpha 1 |
1631 |
0.56 |
| chr13_36048764_36048946 | 0.29 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
1977 |
0.3 |
| chrX_117481980_117482135 | 0.29 |
WDR44 |
WD repeat domain 44 |
1951 |
0.37 |
| chr16_15951360_15951580 | 0.29 |
MYH11 |
myosin, heavy chain 11, smooth muscle |
580 |
0.71 |
| chr2_162166890_162167041 | 0.29 |
PSMD14 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
1854 |
0.36 |
| chr13_45147177_45147503 | 0.29 |
TSC22D1 |
TSC22 domain family, member 1 |
3052 |
0.35 |
| chr18_56244732_56244930 | 0.29 |
RP11-126O1.2 |
|
20529 |
0.14 |
| chr5_146859281_146859432 | 0.28 |
DPYSL3 |
dihydropyrimidinase-like 3 |
25527 |
0.24 |
| chr10_17273946_17274204 | 0.28 |
VIM |
vimentin |
1467 |
0.3 |
| chr4_177711391_177711542 | 0.28 |
VEGFC |
vascular endothelial growth factor C |
2415 |
0.44 |
| chr8_77320139_77320373 | 0.28 |
ENSG00000222231 |
. |
142289 |
0.05 |
| chr14_91478586_91478737 | 0.27 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
47817 |
0.13 |
| chr7_130590792_130591004 | 0.27 |
ENSG00000226380 |
. |
28600 |
0.21 |
| chr2_106260790_106261225 | 0.27 |
NCK2 |
NCK adaptor protein 2 |
100347 |
0.08 |
| chr3_21955222_21955373 | 0.27 |
ENSG00000221384 |
. |
84067 |
0.11 |
| chr9_91791725_91791876 | 0.27 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
1882 |
0.43 |
| chr11_125920446_125920597 | 0.27 |
CDON |
cell adhesion associated, oncogene regulated |
12185 |
0.2 |
| chr5_124078453_124078604 | 0.27 |
ZNF608 |
zinc finger protein 608 |
1686 |
0.26 |
| chr4_21769993_21770244 | 0.27 |
KCNIP4 |
Kv channel interacting protein 4 |
70721 |
0.12 |
| chr9_25677269_25678110 | 0.27 |
TUSC1 |
tumor suppressor candidate 1 |
1167 |
0.68 |
| chr5_92919252_92919403 | 0.27 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
284 |
0.9 |
| chr19_52956436_52957233 | 0.27 |
ZNF578 |
zinc finger protein 578 |
5 |
0.97 |
| chr15_99789888_99790153 | 0.27 |
TTC23 |
tetratricopeptide repeat domain 23 |
205 |
0.9 |
| chr9_140524469_140524620 | 0.27 |
EHMT1 |
euchromatic histone-lysine N-methyltransferase 1 |
11090 |
0.13 |
| chr3_148897296_148897508 | 0.27 |
CP |
ceruloplasmin (ferroxidase) |
6 |
0.98 |
| chr16_73096665_73097349 | 0.26 |
ZFHX3 |
zinc finger homeobox 3 |
3410 |
0.3 |
| chr18_51886495_51886646 | 0.26 |
C18orf54 |
chromosome 18 open reading frame 54 |
1615 |
0.37 |
| chr2_112195055_112195206 | 0.26 |
ENSG00000266139 |
. |
116462 |
0.07 |
| chr3_122201810_122202029 | 0.26 |
KPNA1 |
karyopherin alpha 1 (importin alpha 5) |
31470 |
0.14 |
| chr16_51188825_51189552 | 0.26 |
SALL1 |
spalt-like transcription factor 1 |
4002 |
0.28 |
| chr6_151562009_151562901 | 0.25 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
946 |
0.51 |
| chr6_27722212_27722363 | 0.25 |
HIST1H2BL |
histone cluster 1, H2bl |
53422 |
0.06 |
| chr10_86184623_86185188 | 0.25 |
CCSER2 |
coiled-coil serine-rich protein 2 |
210 |
0.97 |
| chr11_10428526_10428677 | 0.25 |
ENSG00000221574 |
. |
7737 |
0.21 |
| chr17_57449730_57449881 | 0.25 |
ENSG00000263857 |
. |
6361 |
0.19 |
| chr7_134539898_134540267 | 0.25 |
CALD1 |
caldesmon 1 |
11447 |
0.28 |
| chr6_56996023_56996174 | 0.25 |
RP11-203B9.4 |
|
39749 |
0.13 |
| chr1_164529707_164529858 | 0.25 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
722 |
0.78 |
| chr18_53090404_53090609 | 0.25 |
TCF4 |
transcription factor 4 |
763 |
0.7 |
| chr8_29027351_29027502 | 0.25 |
ENSG00000264328 |
. |
27216 |
0.15 |
| chr5_179047232_179047712 | 0.25 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
488 |
0.71 |
| chr3_154517752_154517903 | 0.25 |
MME |
membrane metallo-endopeptidase |
224086 |
0.02 |
| chr2_216283516_216283667 | 0.25 |
FN1 |
fibronectin 1 |
17199 |
0.2 |
| chr3_170302671_170302921 | 0.25 |
SLC7A14 |
solute carrier family 7, member 14 |
1067 |
0.64 |
| chrX_151039282_151039625 | 0.25 |
RP11-366F6.2 |
|
40218 |
0.13 |
| chr2_87812694_87812845 | 0.25 |
RP11-1399P15.1 |
|
35216 |
0.22 |
| chr18_64307786_64307937 | 0.25 |
ENSG00000221536 |
. |
12877 |
0.26 |
| chr12_32261156_32261307 | 0.25 |
RP11-843B15.2 |
|
769 |
0.51 |
| chr3_9945019_9945243 | 0.25 |
IL17RE |
interleukin 17 receptor E |
565 |
0.54 |
| chr12_20525245_20525493 | 0.24 |
RP11-284H19.1 |
|
2173 |
0.35 |
| chr17_59478534_59478762 | 0.24 |
RP11-332H18.5 |
|
1044 |
0.31 |
| chr5_169407172_169407407 | 0.24 |
FAM196B |
family with sequence similarity 196, member B |
455 |
0.87 |
| chr2_145201691_145201842 | 0.24 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
13629 |
0.27 |
| chr4_169951673_169951824 | 0.24 |
RP11-483A20.3 |
|
20167 |
0.17 |
| chr3_23245710_23245894 | 0.24 |
UBE2E2 |
ubiquitin-conjugating enzyme E2E 2 |
1018 |
0.51 |
| chr10_112853696_112853847 | 0.24 |
ADRA2A |
adrenoceptor alpha 2A |
16981 |
0.25 |
| chr4_169616116_169616373 | 0.24 |
ENSG00000206613 |
. |
8377 |
0.21 |
| chr2_197852125_197852688 | 0.24 |
ANKRD44 |
ankyrin repeat domain 44 |
12820 |
0.25 |
| chrX_130962862_130963372 | 0.24 |
ENSG00000200587 |
. |
109867 |
0.07 |
| chr14_69657871_69659182 | 0.24 |
EXD2 |
exonuclease 3'-5' domain containing 2 |
3 |
0.98 |
| chrY_7395560_7395750 | 0.24 |
ENSG00000252472 |
. |
104456 |
0.07 |
| chr7_93552513_93552820 | 0.24 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
1655 |
0.33 |
| chr7_72933105_72933537 | 0.24 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
3281 |
0.19 |
| chr12_117096463_117096618 | 0.24 |
C12orf49 |
chromosome 12 open reading frame 49 |
61392 |
0.12 |
| chr19_17559208_17559695 | 0.23 |
TMEM221 |
transmembrane protein 221 |
75 |
0.93 |
| chr12_5019229_5019602 | 0.23 |
KCNA1 |
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) |
344 |
0.92 |
| chr1_101359369_101359680 | 0.23 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
696 |
0.48 |
| chr16_83847696_83847847 | 0.23 |
HSBP1 |
heat shock factor binding protein 1 |
6177 |
0.18 |
| chrX_133509784_133510099 | 0.23 |
PHF6 |
PHD finger protein 6 |
2538 |
0.28 |
| chr8_99258580_99258731 | 0.23 |
NIPAL2 |
NIPA-like domain containing 2 |
47909 |
0.13 |
| chr17_9264144_9264295 | 0.23 |
RP11-85B7.4 |
|
77728 |
0.09 |
| chr3_58067974_58068125 | 0.23 |
FLNB |
filamin B, beta |
3836 |
0.31 |
| chr8_116678925_116679076 | 0.23 |
TRPS1 |
trichorhinophalangeal syndrome I |
1208 |
0.65 |
| chr10_63422698_63423173 | 0.23 |
C10orf107 |
chromosome 10 open reading frame 107 |
180 |
0.96 |
| chr10_69022435_69022586 | 0.23 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
82309 |
0.11 |
| chr16_85038693_85039085 | 0.23 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
273 |
0.93 |
| chr7_23514811_23514962 | 0.23 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
4800 |
0.19 |
| chr2_36473952_36474103 | 0.23 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
109042 |
0.07 |
| chr17_57505253_57505417 | 0.22 |
RP11-567L7.5 |
|
24671 |
0.18 |
| chr7_44922298_44922675 | 0.22 |
ENSG00000264326 |
. |
1087 |
0.34 |
| chr10_21804577_21804741 | 0.22 |
SKIDA1 |
SKI/DACH domain containing 1 |
2189 |
0.22 |
| chr4_122926968_122927132 | 0.22 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
54141 |
0.14 |
| chr3_64253305_64253544 | 0.22 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
231 |
0.95 |
| chr7_19145557_19145708 | 0.22 |
AC003986.6 |
|
6465 |
0.17 |
| chr22_46476561_46476775 | 0.22 |
FLJ27365 |
hsa-mir-4763 |
476 |
0.66 |
| chr1_101312973_101313135 | 0.22 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
47166 |
0.12 |
| chr5_111083741_111083964 | 0.22 |
NREP |
neuronal regeneration related protein |
8096 |
0.21 |
| chr1_225117686_225118141 | 0.22 |
DNAH14 |
dynein, axonemal, heavy chain 14 |
529 |
0.87 |
| chr21_41421632_41421805 | 0.22 |
ENSG00000263973 |
. |
162640 |
0.04 |
| chr6_133240538_133240737 | 0.22 |
ENSG00000200534 |
. |
102279 |
0.06 |
| chr6_85468375_85468673 | 0.22 |
TBX18 |
T-box 18 |
4549 |
0.33 |
| chr6_46816549_46816814 | 0.22 |
GPR116 |
G protein-coupled receptor 116 |
22921 |
0.18 |
| chr12_27164586_27164737 | 0.22 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
1286 |
0.39 |
| chr16_89392054_89392556 | 0.22 |
AC137932.6 |
|
2673 |
0.19 |
| chr6_165341863_165342014 | 0.22 |
C6orf118 |
chromosome 6 open reading frame 118 |
380404 |
0.01 |
| chr6_19838575_19839711 | 0.22 |
RP1-167F1.2 |
|
168 |
0.95 |
| chr9_10208018_10208266 | 0.22 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
174352 |
0.04 |
| chr14_61910692_61910843 | 0.21 |
PRKCH |
protein kinase C, eta |
1491 |
0.5 |
| chr2_43695032_43695183 | 0.21 |
ENSG00000252804 |
. |
59661 |
0.14 |
| chr8_68864615_68865211 | 0.21 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
560 |
0.86 |
| chr14_57273831_57273982 | 0.21 |
OTX2 |
orthodenticle homeobox 2 |
1525 |
0.35 |
| chr15_98065206_98065387 | 0.21 |
ENSG00000251729 |
. |
49814 |
0.19 |
| chr19_37903473_37903682 | 0.21 |
ZNF569 |
zinc finger protein 569 |
32284 |
0.12 |
| chr13_99630412_99630628 | 0.21 |
DOCK9 |
dedicator of cytokinesis 9 |
182 |
0.96 |
| chr9_35341457_35341792 | 0.21 |
AL160274.1 |
HCG17281; PRO0038; Uncharacterized protein |
19638 |
0.2 |
| chr1_182992978_182993852 | 0.21 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
820 |
0.64 |
| chr10_101419068_101420090 | 0.21 |
ENTPD7 |
ectonucleoside triphosphate diphosphohydrolase 7 |
316 |
0.86 |
| chr8_77317816_77317967 | 0.21 |
ENSG00000222231 |
. |
139924 |
0.05 |
| chr2_225751095_225751267 | 0.21 |
DOCK10 |
dedicator of cytokinesis 10 |
60601 |
0.16 |
| chr11_108767349_108767500 | 0.21 |
ENSG00000201243 |
. |
108820 |
0.07 |
| chr18_51886872_51887023 | 0.21 |
C18orf54 |
chromosome 18 open reading frame 54 |
1992 |
0.33 |
| chr16_11458815_11458966 | 0.21 |
RP11-485G7.6 |
|
15712 |
0.09 |
| chr15_30112674_30113382 | 0.21 |
TJP1 |
tight junction protein 1 |
723 |
0.64 |
| chr15_57353772_57353923 | 0.21 |
ENSG00000222586 |
. |
95252 |
0.08 |
| chr6_80340519_80340952 | 0.21 |
SH3BGRL2 |
SH3 domain binding glutamic acid-rich protein like 2 |
265 |
0.95 |
| chr4_53863026_53863177 | 0.21 |
RP11-752D24.2 |
|
51245 |
0.16 |
| chr1_199824347_199824498 | 0.21 |
ENSG00000200139 |
. |
32865 |
0.22 |
| chr10_89629938_89630093 | 0.21 |
KLLN |
killin, p53-regulated DNA replication inhibitor |
6821 |
0.14 |
| chr4_107939121_107939272 | 0.21 |
DKK2 |
dickkopf WNT signaling pathway inhibitor 2 |
18258 |
0.28 |
| chr16_14282323_14282474 | 0.20 |
MKL2 |
MKL/myocardin-like 2 |
1626 |
0.41 |
| chr1_117117499_117117673 | 0.20 |
CD58 |
CD58 molecule |
3925 |
0.2 |
| chr5_43306439_43306623 | 0.20 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
6795 |
0.21 |
| chr7_120631344_120631706 | 0.20 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
1849 |
0.35 |
| chr3_66139685_66140118 | 0.20 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
20616 |
0.25 |
| chr2_161726328_161726479 | 0.20 |
ENSG00000244372 |
. |
7655 |
0.32 |
| chr8_118990990_118991291 | 0.20 |
EXT1 |
exostosin glycosyltransferase 1 |
131513 |
0.06 |
| chr9_18976648_18977105 | 0.20 |
FAM154A |
family with sequence similarity 154, member A |
56310 |
0.1 |
| chr4_146855885_146856036 | 0.20 |
ZNF827 |
zinc finger protein 827 |
3663 |
0.28 |
| chr4_166252557_166253372 | 0.20 |
MSMO1 |
methylsterol monooxygenase 1 |
3885 |
0.25 |
| chr1_182931948_182932121 | 0.20 |
ENSG00000264768 |
. |
3324 |
0.2 |
| chr6_15399360_15399511 | 0.20 |
JARID2 |
jumonji, AT rich interactive domain 2 |
1654 |
0.48 |
| chr4_4764717_4764868 | 0.20 |
MSX1 |
msh homeobox 1 |
96601 |
0.08 |
| chr12_115340794_115340945 | 0.20 |
ENSG00000252459 |
. |
165360 |
0.04 |
| chr2_38295840_38295991 | 0.20 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
1731 |
0.34 |
| chr8_136478308_136478499 | 0.20 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
7453 |
0.25 |
| chr1_192508213_192508364 | 0.20 |
RP5-1011O1.2 |
|
28059 |
0.18 |
| chr6_52514241_52514392 | 0.20 |
RP1-152L7.5 |
|
15756 |
0.19 |
| chr5_145827885_145828143 | 0.20 |
TCERG1 |
transcription elongation regulator 1 |
1119 |
0.56 |
| chr2_201308521_201308672 | 0.20 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
3224 |
0.27 |
| chr5_102203960_102204196 | 0.20 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
2258 |
0.46 |
| chr2_228028292_228028853 | 0.20 |
COL4A4 |
collagen, type IV, alpha 4 |
257 |
0.8 |
| chr18_22907743_22907894 | 0.20 |
ZNF521 |
zinc finger protein 521 |
23339 |
0.23 |
| chr7_80270291_80270524 | 0.20 |
CD36 |
CD36 molecule (thrombospondin receptor) |
2415 |
0.44 |
| chr1_167298272_167298450 | 0.20 |
POU2F1 |
POU class 2 homeobox 1 |
80 |
0.98 |
| chr6_74423009_74423160 | 0.20 |
RP11-553A21.3 |
|
17230 |
0.19 |
| chr3_190580567_190581193 | 0.20 |
GMNC |
geminin coiled-coil domain containing |
476 |
0.9 |
| chr4_152021608_152021943 | 0.20 |
RPS3A |
ribosomal protein S3A |
750 |
0.52 |
| chr10_36739274_36739523 | 0.20 |
ENSG00000237002 |
. |
11809 |
0.29 |
| chr18_53253489_53253863 | 0.20 |
TCF4 |
transcription factor 4 |
326 |
0.93 |
| chr9_21991792_21991943 | 0.20 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
2543 |
0.19 |
| chr13_43843656_43843899 | 0.20 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
91436 |
0.1 |
| chr12_50608832_50609096 | 0.20 |
RP3-405J10.4 |
|
4423 |
0.14 |
| chr11_78919156_78919307 | 0.19 |
TENM4 |
teneurin transmembrane protein 4 |
7723 |
0.32 |
| chr3_181432212_181432363 | 0.19 |
SOX2 |
SRY (sex determining region Y)-box 2 |
2565 |
0.33 |
| chr6_158080284_158080435 | 0.19 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
66318 |
0.12 |
| chr2_98281303_98281454 | 0.19 |
ENSG00000201806 |
. |
184 |
0.78 |
| chr13_50194281_50194827 | 0.19 |
ARL11 |
ADP-ribosylation factor-like 11 |
7881 |
0.21 |
| chr5_179048814_179048995 | 0.19 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
392 |
0.78 |
| chr21_40985164_40985421 | 0.19 |
C21orf88 |
chromosome 21 open reading frame 88 |
544 |
0.78 |
| chr1_52340451_52340602 | 0.19 |
NRD1 |
nardilysin (N-arginine dibasic convertase) |
3165 |
0.18 |
| chr9_130854010_130854186 | 0.19 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
355 |
0.73 |
| chr12_13353569_13354032 | 0.19 |
EMP1 |
epithelial membrane protein 1 |
4080 |
0.3 |
| chr2_210390985_210391683 | 0.19 |
MAP2 |
microtubule-associated protein 2 |
52705 |
0.18 |
| chr2_191199116_191199267 | 0.19 |
INPP1 |
inositol polyphosphate-1-phosphatase |
9005 |
0.21 |
| chr13_60737053_60737204 | 0.19 |
DIAPH3 |
diaphanous-related formin 3 |
772 |
0.67 |
| chr6_124127968_124128119 | 0.19 |
NKAIN2 |
Na+/K+ transporting ATPase interacting 2 |
2697 |
0.32 |
| chr2_202903591_202903816 | 0.19 |
FZD7 |
frizzled family receptor 7 |
4393 |
0.2 |
| chr2_14633353_14633613 | 0.19 |
FAM84A |
family with sequence similarity 84, member A |
139327 |
0.05 |
| chr7_39649759_39649910 | 0.19 |
AC004837.4 |
|
55 |
0.97 |
| chr2_20927770_20927921 | 0.19 |
GDF7 |
growth differentiation factor 7 |
61421 |
0.12 |
| chr14_45433796_45433989 | 0.19 |
FAM179B |
family with sequence similarity 179, member B |
2450 |
0.23 |
| chr5_95069419_95069570 | 0.19 |
CTD-2154I11.2 |
|
1495 |
0.34 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.3 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.3 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.1 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.5 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.4 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.0 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 0.1 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.2 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0032071 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0044557 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.0 | GO:0072172 | ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:2000272 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0045844 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.3 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0045655 | regulation of monocyte differentiation(GO:0045655) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.3 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |