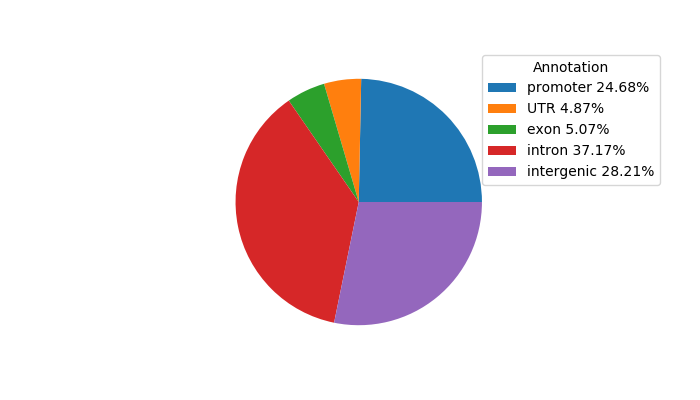
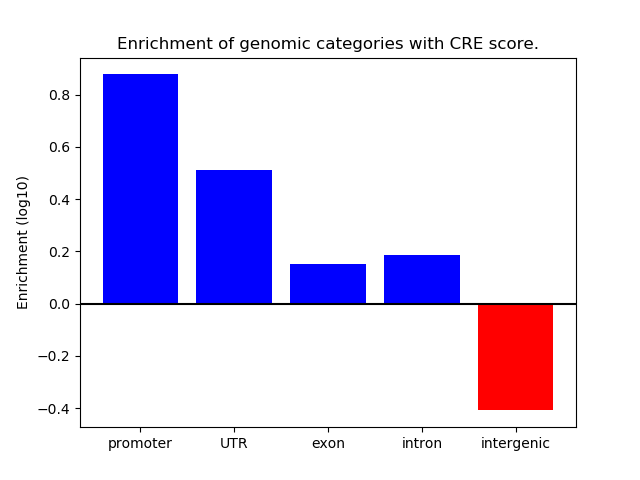
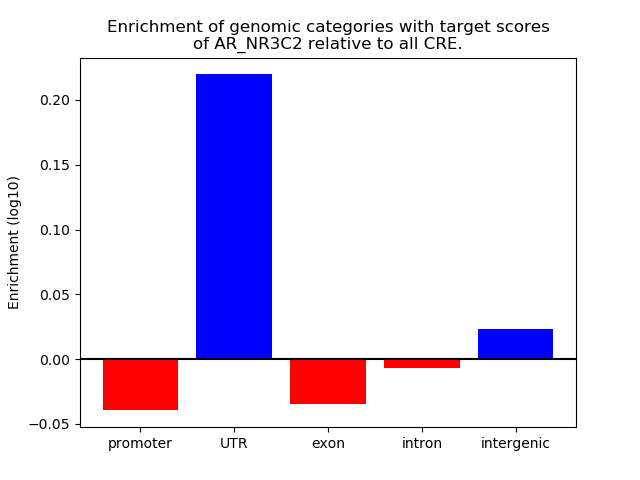
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for AR_NR3C2
Z-value: 0.87
Transcription factors associated with AR_NR3C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
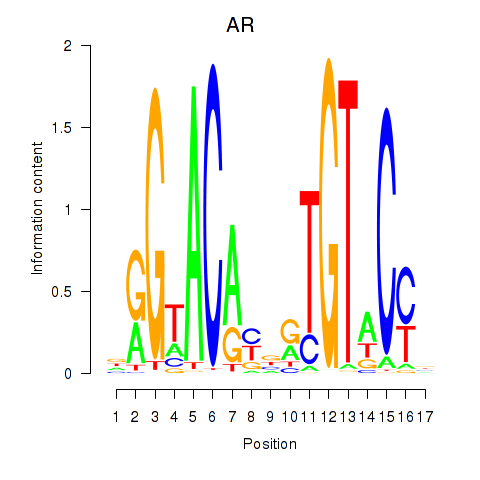
AR
|
ENSG00000169083.11 | androgen receptor |
|
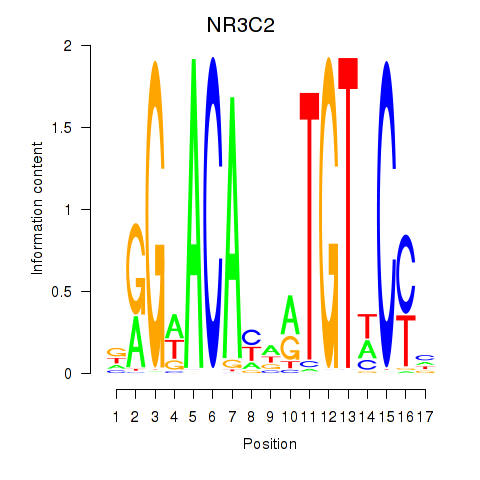
NR3C2
|
ENSG00000151623.10 | nuclear receptor subfamily 3 group C member 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_66766516_66766697 | AR | 1620 | 0.570375 | -0.65 | 5.9e-02 | Click! |
| chrX_66952827_66952978 | AR | 164171 | 0.040013 | 0.63 | 6.8e-02 | Click! |
| chrX_66763846_66764061 | AR | 512 | 0.891958 | -0.61 | 7.8e-02 | Click! |
| chrX_67000951_67001203 | AR | 212346 | 0.025037 | -0.52 | 1.5e-01 | Click! |
| chrX_66767815_66768085 | AR | 2964 | 0.426121 | -0.52 | 1.5e-01 | Click! |
| chr4_149367147_149367363 | NR3C2 | 1405 | 0.591234 | 0.24 | 5.3e-01 | Click! |
| chr4_149363777_149364242 | NR3C2 | 337 | 0.938175 | -0.19 | 6.2e-01 | Click! |
| chr4_149364707_149365251 | NR3C2 | 871 | 0.750626 | 0.15 | 6.9e-01 | Click! |
| chr4_149368732_149368898 | NR3C2 | 2965 | 0.401266 | -0.14 | 7.2e-01 | Click! |
| chr4_149363274_149363592 | NR3C2 | 57 | 0.987392 | -0.12 | 7.5e-01 | Click! |
Activity of the AR_NR3C2 motif across conditions
Conditions sorted by the z-value of the AR_NR3C2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
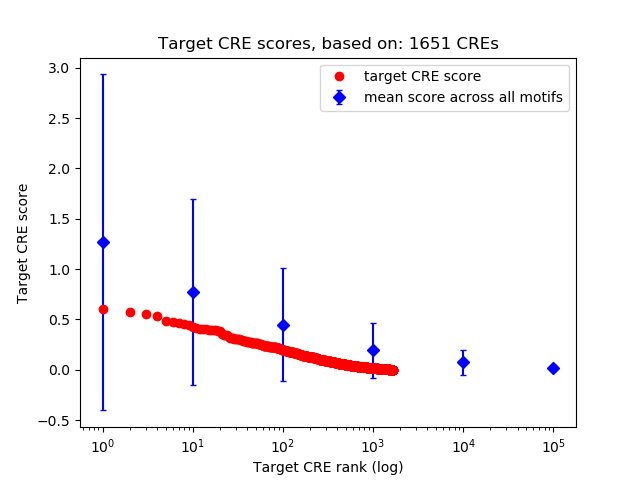
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_118955810_118956058 | 0.61 |
B4GALT4 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
3070 |
0.21 |
| chrX_70760188_70760339 | 0.57 |
OGT |
O-linked N-acetylglucosamine (GlcNAc) transferase |
3399 |
0.24 |
| chr21_46336780_46336931 | 0.55 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
2657 |
0.15 |
| chr1_172715599_172715750 | 0.54 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
87516 |
0.09 |
| chr1_95316335_95316486 | 0.49 |
ENSG00000237416 |
. |
4572 |
0.23 |
| chr5_148309668_148309819 | 0.47 |
RP11-44B19.1 |
|
40160 |
0.16 |
| chr6_11381781_11382025 | 0.46 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
629 |
0.82 |
| chr6_35568962_35570321 | 0.45 |
ENSG00000212579 |
. |
49954 |
0.1 |
| chr8_134084504_134084857 | 0.44 |
SLA |
Src-like-adaptor |
12077 |
0.24 |
| chr16_11794621_11794772 | 0.43 |
SNN |
stannin |
32426 |
0.12 |
| chr15_40659564_40659826 | 0.41 |
RP11-64K12.4 |
|
2146 |
0.13 |
| chr12_113416849_113417235 | 0.40 |
OAS2 |
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
697 |
0.66 |
| chr6_154568354_154568864 | 0.40 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
| chr11_114186668_114186999 | 0.40 |
NNMT |
nicotinamide N-methyltransferase |
17960 |
0.18 |
| chr17_79031323_79031474 | 0.40 |
BAIAP2 |
BAI1-associated protein 2 |
17 |
0.97 |
| chr12_51713121_51713481 | 0.40 |
BIN2 |
bridging integrator 2 |
4598 |
0.18 |
| chr1_10857132_10857283 | 0.40 |
CASZ1 |
castor zinc finger 1 |
500 |
0.85 |
| chr13_46959234_46959385 | 0.39 |
KIAA0226L |
KIAA0226-like |
2008 |
0.32 |
| chr15_86077505_86077656 | 0.39 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
9691 |
0.21 |
| chr6_13448559_13448819 | 0.38 |
AL583828.1 |
|
37726 |
0.13 |
| chr6_36281244_36281395 | 0.36 |
C6orf222 |
chromosome 6 open reading frame 222 |
23343 |
0.15 |
| chr13_111213122_111213273 | 0.35 |
RAB20 |
RAB20, member RAS oncogene family |
883 |
0.64 |
| chr6_42013548_42014157 | 0.35 |
CCND3 |
cyclin D3 |
2572 |
0.23 |
| chr19_16256933_16257251 | 0.34 |
HSH2D |
hematopoietic SH2 domain containing |
2546 |
0.18 |
| chr1_112032205_112032457 | 0.33 |
ADORA3 |
adenosine A3 receptor |
47 |
0.91 |
| chr12_52409491_52409642 | 0.32 |
GRASP |
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
4599 |
0.14 |
| chr12_26441132_26441283 | 0.32 |
RP11-283G6.5 |
|
16354 |
0.18 |
| chr12_40551802_40551953 | 0.32 |
LRRK2 |
leucine-rich repeat kinase 2 |
38669 |
0.17 |
| chr6_25006652_25006894 | 0.31 |
ENSG00000244618 |
. |
24737 |
0.17 |
| chr17_80255361_80255512 | 0.31 |
CD7 |
CD7 molecule |
19992 |
0.09 |
| chr15_63891011_63891162 | 0.31 |
FBXL22 |
F-box and leucine-rich repeat protein 22 |
1185 |
0.42 |
| chr1_150540056_150540997 | 0.30 |
ENSG00000264508 |
. |
1190 |
0.22 |
| chr2_99439255_99439699 | 0.30 |
ENSG00000238830 |
. |
17722 |
0.19 |
| chr11_113981757_113982173 | 0.30 |
ENSG00000221112 |
. |
24314 |
0.2 |
| chr12_53985901_53986052 | 0.30 |
ATF7 |
activating transcription factor 7 |
8829 |
0.15 |
| chr7_150147431_150147649 | 0.29 |
GIMAP8 |
GTPase, IMAP family member 8 |
178 |
0.94 |
| chr15_55547653_55548107 | 0.29 |
RAB27A |
RAB27A, member RAS oncogene family |
6647 |
0.22 |
| chr4_86953999_86954560 | 0.29 |
RP13-514E23.1 |
|
19252 |
0.19 |
| chr6_6916042_6916314 | 0.28 |
ENSG00000240936 |
. |
22952 |
0.26 |
| chr2_109605420_109605703 | 0.28 |
EDAR |
ectodysplasin A receptor |
164 |
0.97 |
| chr8_120126680_120126831 | 0.28 |
RP11-278I4.2 |
|
45734 |
0.15 |
| chr2_37817410_37817581 | 0.28 |
AC006369.2 |
|
9784 |
0.26 |
| chr3_73085691_73085907 | 0.28 |
ENSG00000238959 |
. |
8840 |
0.16 |
| chr11_46363477_46363864 | 0.28 |
DGKZ |
diacylglycerol kinase, zeta |
3147 |
0.2 |
| chrX_118710010_118710161 | 0.27 |
UBE2A |
ubiquitin-conjugating enzyme E2A |
1445 |
0.35 |
| chrX_53100883_53101580 | 0.27 |
TSPYL2 |
TSPY-like 2 |
10318 |
0.18 |
| chr3_81873946_81874241 | 0.27 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
62781 |
0.16 |
| chr1_95018005_95018156 | 0.27 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
10724 |
0.29 |
| chr11_122655510_122655661 | 0.26 |
CRTAM |
cytotoxic and regulatory T cell molecule |
53623 |
0.12 |
| chr6_6750463_6750647 | 0.26 |
LY86-AS1 |
LY86 antisense RNA 1 |
127551 |
0.05 |
| chr12_106178721_106178872 | 0.26 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
299015 |
0.01 |
| chr12_728674_728825 | 0.26 |
NINJ2 |
ninjurin 2 |
9165 |
0.14 |
| chr1_84899098_84899702 | 0.25 |
DNASE2B |
deoxyribonuclease II beta |
25487 |
0.17 |
| chr11_65059695_65060120 | 0.25 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
9929 |
0.12 |
| chr17_64672266_64672417 | 0.25 |
AC006947.1 |
|
149 |
0.96 |
| chr9_95824320_95824471 | 0.25 |
SUSD3 |
sushi domain containing 3 |
3334 |
0.22 |
| chr4_57570517_57570806 | 0.25 |
HOPX |
HOP homeobox |
22596 |
0.2 |
| chr9_119134239_119134390 | 0.25 |
ENSG00000216182 |
. |
21641 |
0.17 |
| chr1_244512207_244512358 | 0.24 |
C1orf100 |
chromosome 1 open reading frame 100 |
3655 |
0.3 |
| chr19_2736707_2736858 | 0.24 |
SLC39A3 |
solute carrier family 39 (zinc transporter), member 3 |
2691 |
0.14 |
| chr13_49234211_49234362 | 0.24 |
CYSLTR2 |
cysteinyl leukotriene receptor 2 |
46665 |
0.17 |
| chr19_8261540_8261691 | 0.24 |
CERS4 |
ceramide synthase 4 |
10005 |
0.16 |
| chr3_105595835_105595986 | 0.24 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
7514 |
0.34 |
| chr5_156731259_156731410 | 0.24 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
6711 |
0.16 |
| chr9_117691491_117691642 | 0.24 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
1131 |
0.63 |
| chr2_71772491_71772642 | 0.24 |
DYSF |
dysferlin |
78734 |
0.11 |
| chr6_35265630_35266434 | 0.24 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
403 |
0.84 |
| chr13_91068921_91069072 | 0.24 |
ENSG00000207858 |
. |
185560 |
0.03 |
| chr3_13920477_13920889 | 0.23 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
935 |
0.64 |
| chr11_72437407_72437702 | 0.23 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
619 |
0.62 |
| chr9_117381662_117382014 | 0.23 |
ENSG00000272241 |
. |
3011 |
0.2 |
| chr14_55799126_55799413 | 0.23 |
RP11-665C16.5 |
|
6950 |
0.25 |
| chr2_112221457_112221687 | 0.23 |
ENSG00000266139 |
. |
142904 |
0.05 |
| chr17_81011527_81011678 | 0.23 |
B3GNTL1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
1916 |
0.37 |
| chr11_65407442_65407916 | 0.23 |
SIPA1 |
signal-induced proliferation-associated 1 |
87 |
0.92 |
| chr18_45856570_45856721 | 0.23 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
8117 |
0.26 |
| chr10_12647585_12647797 | 0.23 |
ENSG00000263584 |
. |
26939 |
0.23 |
| chr15_38944224_38944375 | 0.22 |
C15orf53 |
chromosome 15 open reading frame 53 |
44500 |
0.17 |
| chr3_12185604_12185755 | 0.22 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
15172 |
0.26 |
| chr1_171632627_171632778 | 0.22 |
MYOC |
myocilin, trabecular meshwork inducible glucocorticoid response |
10879 |
0.22 |
| chr12_71167942_71168093 | 0.22 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
14745 |
0.28 |
| chr4_148703706_148703905 | 0.22 |
ENSG00000264274 |
. |
59 |
0.98 |
| chr18_34021681_34021832 | 0.22 |
FHOD3 |
formin homology 2 domain containing 3 |
102758 |
0.08 |
| chr15_68591382_68591806 | 0.22 |
FEM1B |
fem-1 homolog b (C. elegans) |
9044 |
0.21 |
| chr4_38320095_38320594 | 0.22 |
ENSG00000221495 |
. |
80110 |
0.11 |
| chr5_14594534_14594878 | 0.22 |
FAM105A |
family with sequence similarity 105, member A |
12822 |
0.24 |
| chr20_43810738_43810889 | 0.22 |
PI3 |
peptidase inhibitor 3, skin-derived |
7296 |
0.15 |
| chr4_57547872_57548147 | 0.22 |
HOPX |
HOP homeobox |
56 |
0.98 |
| chr19_47660903_47661882 | 0.22 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
27240 |
0.13 |
| chr5_114762279_114762430 | 0.21 |
FEM1C |
fem-1 homolog c (C. elegans) |
118237 |
0.05 |
| chr22_50969510_50970103 | 0.21 |
ODF3B |
outer dense fiber of sperm tails 3B |
700 |
0.36 |
| chr3_1765848_1765999 | 0.21 |
ENSG00000223040 |
. |
364972 |
0.01 |
| chr1_59395732_59395883 | 0.21 |
JUN |
jun proto-oncogene |
146022 |
0.04 |
| chr3_53198863_53199290 | 0.21 |
PRKCD |
protein kinase C, delta |
69 |
0.97 |
| chr1_167586551_167586989 | 0.21 |
RCSD1 |
RCSD domain containing 1 |
12560 |
0.19 |
| chr3_156534921_156535559 | 0.20 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
8849 |
0.29 |
| chr13_48809687_48810120 | 0.20 |
ITM2B |
integral membrane protein 2B |
2564 |
0.37 |
| chr3_141866549_141866700 | 0.20 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
1678 |
0.34 |
| chrX_153209580_153209767 | 0.20 |
RENBP |
renin binding protein |
466 |
0.64 |
| chr12_98864880_98865031 | 0.20 |
RP11-181C3.1 |
Uncharacterized protein |
32678 |
0.13 |
| chr2_206633742_206633893 | 0.20 |
AC007362.3 |
|
5087 |
0.3 |
| chr6_160241928_160242222 | 0.19 |
PNLDC1 |
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
20769 |
0.12 |
| chr1_159796420_159796571 | 0.19 |
SLAMF8 |
SLAM family member 8 |
45 |
0.96 |
| chr5_159214342_159214493 | 0.19 |
ADRA1B |
adrenoceptor alpha 1B |
129373 |
0.05 |
| chr4_149364707_149365251 | 0.19 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
871 |
0.75 |
| chr11_919324_919475 | 0.19 |
CHID1 |
chitinase domain containing 1 |
4341 |
0.12 |
| chr21_16816295_16816446 | 0.19 |
ENSG00000212564 |
. |
170232 |
0.04 |
| chr8_145806326_145806590 | 0.19 |
CTD-2517M22.9 |
|
2966 |
0.14 |
| chrX_131159258_131159409 | 0.19 |
MST4 |
Serine/threonine-protein kinase MST4 |
1697 |
0.42 |
| chr2_144577318_144577469 | 0.19 |
CTD-2252P21.1 |
|
43751 |
0.16 |
| chr14_45722560_45723019 | 0.19 |
MIS18BP1 |
MIS18 binding protein 1 |
184 |
0.95 |
| chr3_138839315_138839466 | 0.19 |
MRPS22 |
mitochondrial ribosomal protein S22 |
10144 |
0.24 |
| chr13_42294870_42295021 | 0.19 |
ENSG00000241406 |
. |
86868 |
0.09 |
| chr6_111391598_111391749 | 0.18 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
17108 |
0.16 |
| chr3_195641693_195641881 | 0.18 |
AC124944.3 |
|
2822 |
0.17 |
| chr16_22217341_22217492 | 0.18 |
EEF2K |
eukaryotic elongation factor-2 kinase |
187 |
0.95 |
| chr9_2025279_2025430 | 0.18 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
3409 |
0.31 |
| chr5_118606701_118606876 | 0.18 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2339 |
0.29 |
| chr14_100536973_100537221 | 0.18 |
CTD-2376I20.1 |
|
4118 |
0.17 |
| chr19_2281673_2281974 | 0.18 |
C19orf35 |
chromosome 19 open reading frame 35 |
352 |
0.73 |
| chr13_99955996_99956281 | 0.18 |
GPR183 |
G protein-coupled receptor 183 |
3521 |
0.25 |
| chr2_201991504_201992128 | 0.18 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
2236 |
0.22 |
| chr12_1005549_1005700 | 0.17 |
WNK1 |
WNK lysine deficient protein kinase 1 |
4139 |
0.25 |
| chr10_33633036_33633187 | 0.17 |
NRP1 |
neuropilin 1 |
7921 |
0.28 |
| chr9_86886547_86886817 | 0.17 |
RP11-380F14.2 |
|
6452 |
0.29 |
| chr8_91013758_91014471 | 0.17 |
DECR1 |
2,4-dienoyl CoA reductase 1, mitochondrial |
369 |
0.89 |
| chr8_62602537_62602767 | 0.17 |
ASPH |
aspartate beta-hydroxylase |
244 |
0.94 |
| chr5_135344089_135344240 | 0.17 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
20420 |
0.19 |
| chr2_5835221_5835372 | 0.17 |
SOX11 |
SRY (sex determining region Y)-box 11 |
2497 |
0.27 |
| chr3_187457028_187457694 | 0.17 |
BCL6 |
B-cell CLL/lymphoma 6 |
1629 |
0.41 |
| chr5_141391978_141392551 | 0.17 |
GNPDA1 |
glucosamine-6-phosphate deaminase 1 |
95 |
0.96 |
| chr10_43905439_43905728 | 0.17 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
969 |
0.49 |
| chr3_126074801_126075538 | 0.17 |
KLF15 |
Kruppel-like factor 15 |
1116 |
0.51 |
| chr1_38276768_38276919 | 0.17 |
C1orf122 |
chromosome 1 open reading frame 122 |
2761 |
0.13 |
| chr10_25465646_25465797 | 0.17 |
GPR158-AS1 |
GPR158 antisense RNA 1 |
516 |
0.76 |
| chr2_87785010_87785930 | 0.17 |
RP11-1399P15.1 |
|
7917 |
0.31 |
| chr8_102456764_102457065 | 0.17 |
ENSG00000239211 |
. |
46753 |
0.13 |
| chr1_110166249_110166733 | 0.17 |
AMPD2 |
adenosine monophosphate deaminase 2 |
55 |
0.94 |
| chr10_74014620_74015053 | 0.17 |
DDIT4 |
DNA-damage-inducible transcript 4 |
18842 |
0.13 |
| chr11_114030925_114031331 | 0.16 |
ENSG00000221112 |
. |
73477 |
0.1 |
| chr4_38623548_38623699 | 0.16 |
RP11-617D20.1 |
|
2573 |
0.31 |
| chr16_29755026_29755177 | 0.16 |
AC009133.17 |
|
1467 |
0.21 |
| chr16_66241067_66241218 | 0.16 |
ENSG00000201999 |
. |
94570 |
0.08 |
| chr9_132082698_132083492 | 0.16 |
ENSG00000242281 |
. |
49645 |
0.12 |
| chr1_46073171_46073322 | 0.16 |
NASP |
nuclear autoantigenic sperm protein (histone-binding) |
5595 |
0.17 |
| chr3_116582620_116583104 | 0.16 |
ENSG00000265433 |
. |
13648 |
0.25 |
| chr3_13038459_13038610 | 0.16 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
9998 |
0.27 |
| chr1_92925539_92925737 | 0.16 |
GFI1 |
growth factor independent 1 transcription repressor |
23873 |
0.21 |
| chr20_34031080_34031231 | 0.16 |
GDF5 |
growth differentiation factor 5 |
5132 |
0.12 |
| chr3_134203579_134204194 | 0.15 |
CEP63 |
centrosomal protein 63kDa |
699 |
0.51 |
| chr12_27900729_27900880 | 0.15 |
ENSG00000252585 |
. |
5003 |
0.13 |
| chr11_65185396_65185663 | 0.15 |
ENSG00000245532 |
. |
26400 |
0.09 |
| chr2_169964875_169965026 | 0.15 |
AC007556.3 |
|
7697 |
0.23 |
| chr17_692703_692854 | 0.15 |
RP11-676J12.8 |
|
987 |
0.43 |
| chr4_10021309_10021460 | 0.15 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
1723 |
0.34 |
| chr15_48310054_48310205 | 0.15 |
SLC24A5 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
103040 |
0.07 |
| chr6_86125936_86126087 | 0.15 |
NT5E |
5'-nucleotidase, ecto (CD73) |
33798 |
0.22 |
| chr6_209678_209938 | 0.15 |
DUSP22 |
dual specificity phosphatase 22 |
81822 |
0.11 |
| chr1_1148809_1149188 | 0.15 |
TNFRSF4 |
tumor necrosis factor receptor superfamily, member 4 |
514 |
0.56 |
| chr9_84295213_84295364 | 0.15 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
6995 |
0.27 |
| chr9_92255731_92255882 | 0.15 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
35853 |
0.22 |
| chr6_1578010_1578161 | 0.15 |
FOXC1 |
forkhead box C1 |
32596 |
0.22 |
| chr9_92079076_92079388 | 0.15 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
8364 |
0.24 |
| chr8_77529270_77529421 | 0.15 |
ZFHX4-AS1 |
ZFHX4 antisense RNA 1 |
669 |
0.77 |
| chr11_61276967_61277195 | 0.15 |
LRRC10B |
leucine rich repeat containing 10B |
809 |
0.41 |
| chr14_75807662_75807918 | 0.14 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
60894 |
0.09 |
| chr11_16120543_16120694 | 0.14 |
ENSG00000221556 |
. |
25244 |
0.25 |
| chr2_101664673_101664824 | 0.14 |
ENSG00000265860 |
. |
45283 |
0.11 |
| chr1_216130942_216131093 | 0.14 |
RP5-1111A8.3 |
|
71093 |
0.13 |
| chr2_68959667_68959843 | 0.14 |
ARHGAP25 |
Rho GTPase activating protein 25 |
2158 |
0.4 |
| chr1_145427603_145427863 | 0.14 |
TXNIP |
thioredoxin interacting protein |
10736 |
0.12 |
| chr5_148641029_148641180 | 0.14 |
AFAP1L1 |
actin filament associated protein 1-like 1 |
10330 |
0.13 |
| chr10_43894836_43894987 | 0.14 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
2117 |
0.26 |
| chr1_161494601_161494967 | 0.14 |
HSPA6 |
heat shock 70kDa protein 6 (HSP70B') |
748 |
0.56 |
| chr19_2290395_2291381 | 0.14 |
LINGO3 |
leucine rich repeat and Ig domain containing 3 |
1135 |
0.28 |
| chr10_14643816_14644958 | 0.14 |
FAM107B |
family with sequence similarity 107, member B |
2001 |
0.38 |
| chr6_111926441_111927468 | 0.14 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
120 |
0.96 |
| chr16_66971123_66971274 | 0.14 |
CES2 |
carboxylesterase 2 |
2820 |
0.14 |
| chr16_16308222_16308373 | 0.14 |
RP11-517A5.7 |
|
8587 |
0.15 |
| chr8_131024317_131024468 | 0.14 |
ENSG00000264653 |
. |
3693 |
0.2 |
| chr7_77044826_77045681 | 0.14 |
GSAP |
gamma-secretase activating protein |
69 |
0.98 |
| chr17_28691143_28691377 | 0.13 |
ENSG00000201255 |
. |
3779 |
0.16 |
| chr5_79230796_79230947 | 0.13 |
RP11-168A11.1 |
|
1531 |
0.47 |
| chr3_194592076_194592227 | 0.13 |
FAM43A |
family with sequence similarity 43, member A |
185529 |
0.02 |
| chr4_139230379_139230530 | 0.13 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
66951 |
0.14 |
| chr14_65545748_65545899 | 0.13 |
RP11-840I19.3 |
|
2929 |
0.21 |
| chr2_145129966_145130117 | 0.13 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
39958 |
0.19 |
| chr11_6767215_6767876 | 0.13 |
OR2AG2 |
olfactory receptor, family 2, subfamily AG, member 2 |
22741 |
0.11 |
| chr4_121082_121430 | 0.13 |
ENSG00000211553 |
. |
8986 |
0.22 |
| chr9_33162993_33163393 | 0.13 |
RP11-326F20.5 |
|
3780 |
0.17 |
| chrX_25031023_25031386 | 0.13 |
ARX |
aristaless related homeobox |
2861 |
0.36 |
| chr2_39472655_39472806 | 0.13 |
CDKL4 |
cyclin-dependent kinase-like 4 |
16057 |
0.19 |
| chr4_87764800_87764951 | 0.13 |
SLC10A6 |
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
5541 |
0.27 |
| chr16_87166401_87166552 | 0.13 |
RP11-899L11.3 |
|
83045 |
0.09 |
| chr5_97764803_97764954 | 0.13 |
ENSG00000223053 |
. |
210057 |
0.03 |
| chr10_74089726_74089877 | 0.13 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
14918 |
0.18 |
| chr7_100806219_100806526 | 0.13 |
VGF |
VGF nerve growth factor inducible |
2023 |
0.17 |
| chr2_175528047_175528198 | 0.13 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
19477 |
0.21 |
| chr11_130783512_130783663 | 0.13 |
SNX19 |
sorting nexin 19 |
2757 |
0.4 |
| chr4_24474548_24475165 | 0.13 |
ENSG00000265001 |
. |
43401 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.3 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.2 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.0 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.0 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0071220 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.0 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |