Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
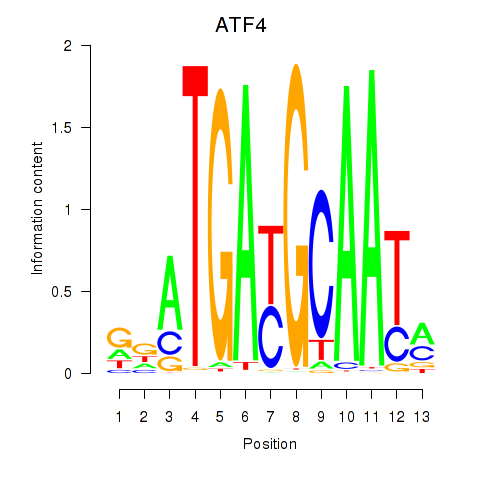
Results for ATF4
Z-value: 0.35
Transcription factors associated with ATF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF4
|
ENSG00000128272.10 | activating transcription factor 4 |
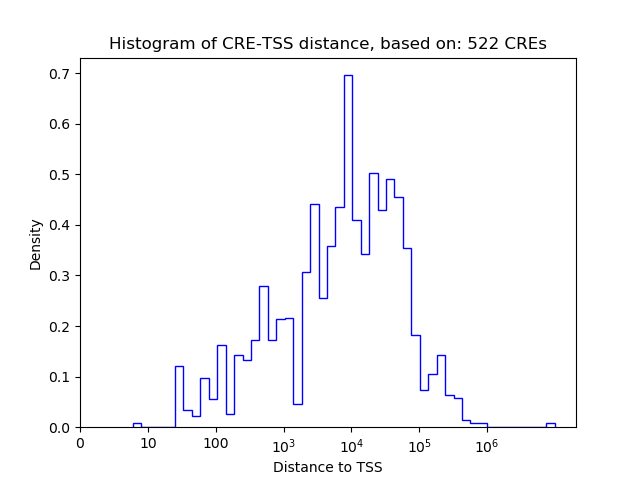
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr22_39916629_39916983 | ATF4 | 237 | 0.900609 | -0.59 | 9.3e-02 | Click! |
| chr22_39915742_39916052 | ATF4 | 197 | 0.920332 | -0.52 | 1.5e-01 | Click! |
| chr22_39917795_39917946 | ATF4 | 1301 | 0.361515 | -0.52 | 1.5e-01 | Click! |
| chr22_39917253_39917786 | ATF4 | 950 | 0.477950 | -0.50 | 1.7e-01 | Click! |
| chr22_39916301_39916487 | ATF4 | 175 | 0.930203 | -0.49 | 1.8e-01 | Click! |
Activity of the ATF4 motif across conditions
Conditions sorted by the z-value of the ATF4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
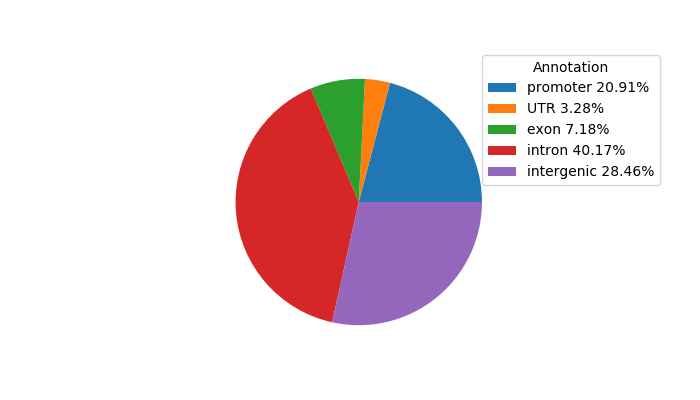
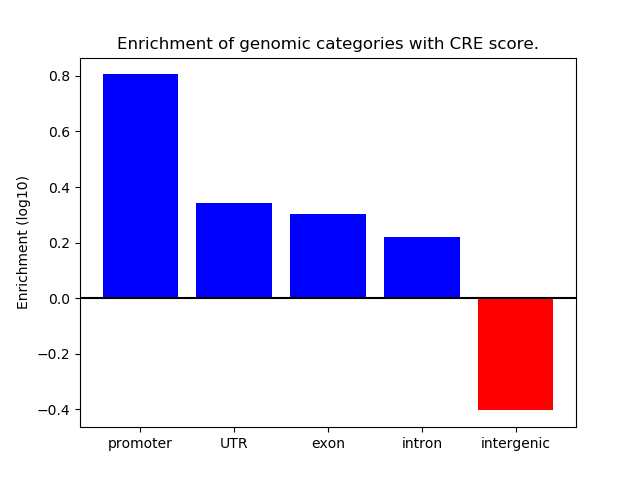
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_50919615_50920637 | 0.14 |
ADM2 |
adrenomedullin 2 |
27 |
0.94 |
| chr5_153691070_153691221 | 0.14 |
ENSG00000221070 |
. |
28541 |
0.16 |
| chr3_5239428_5239587 | 0.12 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
9473 |
0.15 |
| chr1_59966766_59966917 | 0.11 |
FGGY |
FGGY carbohydrate kinase domain containing |
52749 |
0.18 |
| chr20_43124073_43124404 | 0.10 |
SERINC3 |
serine incorporator 3 |
9294 |
0.12 |
| chr13_95133005_95133156 | 0.10 |
DCT |
dopachrome tautomerase |
1144 |
0.56 |
| chr1_234039912_234040198 | 0.09 |
SLC35F3 |
solute carrier family 35, member F3 |
624 |
0.81 |
| chr19_19513882_19514033 | 0.09 |
CTB-184G21.3 |
|
545 |
0.68 |
| chrX_49853365_49853516 | 0.09 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
19284 |
0.14 |
| chr6_88609819_88609970 | 0.09 |
ENSG00000211983 |
. |
52544 |
0.13 |
| chr8_96247980_96248131 | 0.08 |
C8orf37 |
chromosome 8 open reading frame 37 |
33374 |
0.2 |
| chr13_114263460_114263611 | 0.08 |
TFDP1 |
transcription factor Dp-1 |
23794 |
0.17 |
| chr4_185734510_185734661 | 0.08 |
RP11-701P16.2 |
Uncharacterized protein |
188 |
0.91 |
| chr11_20000428_20000721 | 0.08 |
NAV2-AS3 |
NAV2 antisense RNA 3 |
2309 |
0.36 |
| chr11_70262295_70262446 | 0.07 |
CTTN |
cortactin |
3861 |
0.17 |
| chr10_28031252_28032104 | 0.07 |
MKX |
mohawk homeobox |
796 |
0.61 |
| chr1_8278863_8279147 | 0.07 |
ENSG00000200975 |
. |
12348 |
0.21 |
| chr11_70163755_70163906 | 0.07 |
CTA-797E19.1 |
|
2204 |
0.21 |
| chr16_14994956_14995107 | 0.07 |
ENSG00000257264 |
. |
334 |
0.79 |
| chr19_40590982_40591133 | 0.07 |
ZNF780A |
zinc finger protein 780A |
5085 |
0.16 |
| chr9_36144726_36145049 | 0.07 |
GLIPR2 |
GLI pathogenesis-related 2 |
8145 |
0.19 |
| chr5_159510080_159510300 | 0.07 |
PWWP2A |
PWWP domain containing 2A |
36239 |
0.14 |
| chr1_117079785_117079936 | 0.07 |
CD58 |
CD58 molecule |
7352 |
0.18 |
| chr6_121329824_121330111 | 0.07 |
ENSG00000200310 |
. |
322200 |
0.01 |
| chrX_50200272_50200423 | 0.07 |
CCNB3 |
cyclin B3 |
172807 |
0.03 |
| chr11_72546027_72546178 | 0.06 |
FCHSD2 |
FCH and double SH3 domains 2 |
8456 |
0.14 |
| chr12_29588619_29588770 | 0.06 |
OVCH1 |
ovochymase 1 |
8475 |
0.22 |
| chr9_123495329_123495480 | 0.06 |
MEGF9 |
multiple EGF-like-domains 9 |
18656 |
0.21 |
| chr1_207509019_207509170 | 0.06 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
13958 |
0.26 |
| chr5_142373997_142374148 | 0.06 |
ARHGAP26 |
Rho GTPase activating protein 26 |
42689 |
0.2 |
| chr5_52327571_52327722 | 0.06 |
CTD-2175A23.1 |
|
41538 |
0.14 |
| chr2_46769122_46769585 | 0.06 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
343 |
0.61 |
| chr11_7998839_7999271 | 0.06 |
EIF3F |
eukaryotic translation initiation factor 3, subunit F |
7257 |
0.13 |
| chr2_51447556_51447707 | 0.05 |
NRXN1 |
neurexin 1 |
187957 |
0.03 |
| chr9_40632604_40633916 | 0.05 |
SPATA31A3 |
SPATA31 subfamily A, member 3 |
67031 |
0.14 |
| chr10_29916878_29917029 | 0.05 |
SVIL |
supervillin |
6948 |
0.21 |
| chr11_102096966_102097117 | 0.05 |
RP11-864G5.3 |
|
4612 |
0.23 |
| chr11_35195556_35195728 | 0.05 |
CD44 |
CD44 molecule (Indian blood group) |
2476 |
0.25 |
| chr2_17774779_17774930 | 0.05 |
VSNL1 |
visinin-like 1 |
52427 |
0.16 |
| chr6_24922511_24922662 | 0.05 |
FAM65B |
family with sequence similarity 65, member B |
11391 |
0.23 |
| chr12_65131454_65131605 | 0.05 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
7141 |
0.15 |
| chr17_60859401_60859552 | 0.05 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
22124 |
0.2 |
| chr19_54327344_54327551 | 0.05 |
NLRP12 |
NLR family, pyrin domain containing 12 |
124 |
0.86 |
| chr9_41432727_41433091 | 0.05 |
SPATA31A5 |
SPATA31 subfamily A, member 5 |
67770 |
0.13 |
| chr8_36720546_36720697 | 0.05 |
KCNU1 |
potassium channel, subfamily U, member 1 |
65802 |
0.14 |
| chr17_68166405_68166607 | 0.05 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
830 |
0.45 |
| chr20_47368252_47368403 | 0.05 |
ENSG00000251876 |
. |
12342 |
0.27 |
| chr18_3597270_3597421 | 0.05 |
DLGAP1-AS1 |
DLGAP1 antisense RNA 1 |
2892 |
0.21 |
| chr9_39287322_39287607 | 0.05 |
CNTNAP3 |
contactin associated protein-like 3 |
628 |
0.81 |
| chrX_19700900_19701219 | 0.05 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
11943 |
0.3 |
| chr21_44495024_44495938 | 0.05 |
CBS |
cystathionine-beta-synthase |
460 |
0.8 |
| chr9_65576390_65577630 | 0.04 |
SPATA31A7 |
SPATA31 subfamily A, member 7 |
67400 |
0.15 |
| chr20_61303254_61303405 | 0.04 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
4164 |
0.14 |
| chr2_192058733_192058884 | 0.04 |
STAT4 |
signal transducer and activator of transcription 4 |
42486 |
0.15 |
| chr9_39816703_39816854 | 0.04 |
SPATA31A2 |
SPATA31 subfamily A, member 2 |
68197 |
0.13 |
| chr2_242269478_242269629 | 0.04 |
SEPT2 |
septin 2 |
5705 |
0.15 |
| chr4_68303805_68303956 | 0.04 |
ENSG00000206629 |
. |
8408 |
0.29 |
| chr1_158977592_158977743 | 0.04 |
IFI16 |
interferon, gamma-inducible protein 16 |
1118 |
0.53 |
| chr11_7621443_7621674 | 0.04 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
3141 |
0.25 |
| chr2_87782164_87782461 | 0.04 |
RP11-1399P15.1 |
|
4759 |
0.34 |
| chr11_118111552_118111703 | 0.04 |
MPZL3 |
myelin protein zero-like 3 |
11435 |
0.13 |
| chr1_232756314_232756465 | 0.04 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
59085 |
0.14 |
| chr3_39372377_39372528 | 0.04 |
CCR8 |
chemokine (C-C motif) receptor 8 |
1188 |
0.47 |
| chr3_10231782_10231933 | 0.04 |
IRAK2 |
interleukin-1 receptor-associated kinase 2 |
25308 |
0.1 |
| chr19_31807183_31807454 | 0.04 |
AC007796.1 |
|
32469 |
0.2 |
| chr5_53736380_53736531 | 0.04 |
HSPB3 |
heat shock 27kDa protein 3 |
14990 |
0.27 |
| chr15_31727307_31728004 | 0.04 |
KLF13 |
Kruppel-like factor 13 |
69298 |
0.13 |
| chr1_78392239_78392485 | 0.04 |
NEXN |
nexilin (F actin binding protein) |
8549 |
0.18 |
| chr3_111260963_111261300 | 0.04 |
CD96 |
CD96 molecule |
134 |
0.97 |
| chr22_22417650_22417801 | 0.04 |
IGLVV-66 |
immunoglobulin lambda variable (V)-66 (pseudogene) |
2236 |
0.12 |
| chr15_86099150_86099301 | 0.04 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
548 |
0.77 |
| chr2_197071722_197071873 | 0.04 |
ENSG00000239161 |
. |
8249 |
0.2 |
| chr17_29645268_29645505 | 0.04 |
CTD-2370N5.3 |
|
272 |
0.86 |
| chr12_57349520_57349671 | 0.04 |
RDH16 |
retinol dehydrogenase 16 (all-trans) |
2508 |
0.18 |
| chr1_29254208_29254684 | 0.04 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
13355 |
0.18 |
| chr8_1968982_1969133 | 0.04 |
MYOM2 |
myomesin 2 |
24098 |
0.23 |
| chr16_16393606_16393757 | 0.04 |
ENSG00000257381 |
. |
335 |
0.78 |
| chr9_22006120_22006693 | 0.04 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
2546 |
0.22 |
| chr5_54456050_54456451 | 0.03 |
GPX8 |
glutathione peroxidase 8 (putative) |
252 |
0.86 |
| chr15_66662520_66662887 | 0.03 |
TIPIN |
TIMELESS interacting protein |
13649 |
0.11 |
| chr13_46870401_46870552 | 0.03 |
ENSG00000252854 |
. |
46399 |
0.12 |
| chr1_52520143_52521733 | 0.03 |
TXNDC12 |
thioredoxin domain containing 12 (endoplasmic reticulum) |
75 |
0.92 |
| chr14_55769874_55770025 | 0.03 |
FBXO34 |
F-box protein 34 |
31077 |
0.17 |
| chr1_186782436_186782587 | 0.03 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
15574 |
0.29 |
| chr7_98000760_98000949 | 0.03 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
29526 |
0.17 |
| chr10_14799506_14799657 | 0.03 |
FAM107B |
family with sequence similarity 107, member B |
17315 |
0.21 |
| chr11_34393219_34393432 | 0.03 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
13770 |
0.25 |
| chr14_80296454_80296605 | 0.03 |
RP11-242P2.1 |
|
38923 |
0.23 |
| chrX_12974492_12974668 | 0.03 |
TMSB4X |
thymosin beta 4, X-linked |
18647 |
0.19 |
| chr13_30168298_30169638 | 0.03 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
857 |
0.74 |
| chr8_49831086_49831370 | 0.03 |
SNAI2 |
snail family zinc finger 2 |
2760 |
0.41 |
| chr15_55695695_55695873 | 0.03 |
CCPG1 |
cell cycle progression 1 |
4466 |
0.19 |
| chr3_171805158_171805309 | 0.03 |
FNDC3B |
fibronectin type III domain containing 3B |
39531 |
0.2 |
| chr1_160611300_160611550 | 0.03 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
5386 |
0.18 |
| chr10_65227518_65227669 | 0.03 |
JMJD1C |
jumonji domain containing 1C |
1871 |
0.34 |
| chr19_10865922_10866073 | 0.03 |
ENSG00000265879 |
. |
24933 |
0.1 |
| chr7_115912354_115912783 | 0.03 |
TES |
testis derived transcript (3 LIM domains) |
22079 |
0.19 |
| chr20_23238774_23238931 | 0.03 |
NXT1 |
NTF2-like export factor 1 |
92521 |
0.05 |
| chr16_85558650_85558942 | 0.03 |
ENSG00000264203 |
. |
83698 |
0.08 |
| chr17_45743155_45743306 | 0.03 |
KPNB1 |
karyopherin (importin) beta 1 |
2501 |
0.2 |
| chr6_54063055_54063206 | 0.03 |
MLIP |
muscular LMNA-interacting protein |
86822 |
0.09 |
| chr5_126097836_126098030 | 0.03 |
ENSG00000252185 |
. |
6824 |
0.23 |
| chr6_41168130_41168571 | 0.03 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
582 |
0.65 |
| chr1_244816518_244817656 | 0.03 |
DESI2 |
desumoylating isopeptidase 2 |
34 |
0.98 |
| chr12_8608652_8608803 | 0.03 |
CLEC6A |
C-type lectin domain family 6, member A |
205 |
0.93 |
| chr22_17599343_17599494 | 0.03 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
2725 |
0.2 |
| chr5_131795223_131795415 | 0.03 |
ENSG00000202533 |
. |
8520 |
0.13 |
| chr4_74587190_74587341 | 0.03 |
IL8 |
interleukin 8 |
18958 |
0.22 |
| chr8_126590914_126591065 | 0.03 |
ENSG00000266452 |
. |
134182 |
0.05 |
| chr4_15680427_15680704 | 0.03 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
2612 |
0.25 |
| chr17_38498589_38499681 | 0.03 |
CTD-2267D19.2 |
|
253 |
0.69 |
| chr1_164717921_164718072 | 0.03 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
24153 |
0.17 |
| chr9_110310779_110311295 | 0.03 |
KLF4 |
Kruppel-like factor 4 (gut) |
58274 |
0.13 |
| chrX_110339181_110339857 | 0.03 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
69 |
0.99 |
| chr20_19052062_19052213 | 0.03 |
ENSG00000264669 |
. |
54292 |
0.15 |
| chrX_9431281_9431551 | 0.03 |
TBL1X |
transducin (beta)-like 1X-linked |
45 |
0.99 |
| chr10_63995828_63995979 | 0.03 |
RTKN2 |
rhotekin 2 |
119 |
0.98 |
| chr15_51172886_51173037 | 0.03 |
AP4E1 |
adaptor-related protein complex 4, epsilon 1 subunit |
27909 |
0.22 |
| chr18_13464907_13465379 | 0.02 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
129 |
0.93 |
| chr3_58321087_58321238 | 0.02 |
PXK |
PX domain containing serine/threonine kinase |
2129 |
0.32 |
| chr2_54517166_54517317 | 0.02 |
TSPYL6 |
TSPY-like 6 |
33832 |
0.18 |
| chr1_154130279_154130430 | 0.02 |
NUP210L |
nucleoporin 210kDa-like |
2762 |
0.15 |
| chr11_47241983_47242228 | 0.02 |
RP11-17G12.2 |
|
1197 |
0.29 |
| chr9_117657275_117657426 | 0.02 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
35347 |
0.2 |
| chr12_67928012_67928180 | 0.02 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
114022 |
0.07 |
| chr1_43036666_43036817 | 0.02 |
ENSG00000200254 |
. |
2037 |
0.33 |
| chr10_97624941_97625092 | 0.02 |
RP11-429G19.3 |
|
31751 |
0.15 |
| chr9_107826999_107827201 | 0.02 |
ENSG00000201583 |
. |
31937 |
0.22 |
| chr6_163730438_163730589 | 0.02 |
ENSG00000239136 |
. |
82571 |
0.1 |
| chr18_41514870_41515021 | 0.02 |
ENSG00000202250 |
. |
102715 |
0.08 |
| chr20_14985685_14985836 | 0.02 |
ENSG00000199803 |
. |
16545 |
0.2 |
| chr3_62573316_62573467 | 0.02 |
CADPS |
Ca++-dependent secretion activator |
2470 |
0.43 |
| chr2_30695318_30695469 | 0.02 |
LCLAT1 |
lysocardiolipin acyltransferase 1 |
25124 |
0.22 |
| chr9_110377598_110377749 | 0.02 |
ENSG00000202308 |
. |
47854 |
0.17 |
| chr4_40731755_40731906 | 0.02 |
NSUN7 |
NOP2/Sun domain family, member 7 |
20084 |
0.21 |
| chr3_33687128_33687279 | 0.02 |
CLASP2 |
cytoplasmic linker associated protein 2 |
208 |
0.97 |
| chr2_225543043_225543293 | 0.02 |
CUL3 |
cullin 3 |
93058 |
0.09 |
| chr13_40889454_40889605 | 0.02 |
ENSG00000207458 |
. |
88565 |
0.09 |
| chr14_60239555_60239706 | 0.02 |
RTN1 |
reticulon 1 |
98054 |
0.07 |
| chr1_212781685_212782134 | 0.02 |
ATF3 |
activating transcription factor 3 |
103 |
0.96 |
| chr10_90031525_90031822 | 0.02 |
ENSG00000200891 |
. |
277221 |
0.01 |
| chr19_47134901_47135052 | 0.02 |
GNG8 |
guanine nucleotide binding protein (G protein), gamma 8 |
2966 |
0.12 |
| chr16_9166928_9167149 | 0.02 |
C16orf72 |
chromosome 16 open reading frame 72 |
18467 |
0.16 |
| chr8_25058747_25058898 | 0.02 |
DOCK5 |
dedicator of cytokinesis 5 |
16442 |
0.23 |
| chr3_179754024_179754473 | 0.02 |
PEX5L |
peroxisomal biogenesis factor 5-like |
310 |
0.94 |
| chr18_72235350_72235501 | 0.02 |
RP11-231E4.3 |
|
6090 |
0.21 |
| chr14_95991348_95991499 | 0.02 |
ENSG00000252481 |
. |
8543 |
0.14 |
| chr19_1259849_1260000 | 0.02 |
CIRBP |
cold inducible RNA binding protein |
540 |
0.56 |
| chr17_53573866_53574017 | 0.02 |
ENSG00000200107 |
. |
55230 |
0.15 |
| chr15_89181571_89183025 | 0.02 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
114 |
0.96 |
| chr5_139680846_139680997 | 0.02 |
PFDN1 |
prefoldin subunit 1 |
1747 |
0.3 |
| chr2_85972153_85972528 | 0.02 |
ATOH8 |
atonal homolog 8 (Drosophila) |
8677 |
0.15 |
| chr8_61021784_61021935 | 0.02 |
CA8 |
carbonic anhydrase VIII |
172112 |
0.04 |
| chr21_34712478_34712629 | 0.02 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
15275 |
0.17 |
| chr17_4802345_4802927 | 0.02 |
C17orf107 |
chromosome 17 open reading frame 107 |
77 |
0.92 |
| chr12_47617851_47618002 | 0.02 |
PCED1B |
PC-esterase domain containing 1B |
598 |
0.78 |
| chr8_35828319_35828470 | 0.02 |
AC012215.1 |
Uncharacterized protein |
179029 |
0.03 |
| chr3_71115813_71116298 | 0.02 |
FOXP1 |
forkhead box P1 |
1137 |
0.67 |
| chr7_75575266_75575417 | 0.02 |
POR |
P450 (cytochrome) oxidoreductase |
701 |
0.6 |
| chr4_185235485_185235636 | 0.02 |
ENSG00000244512 |
. |
35471 |
0.16 |
| chr16_88271666_88271817 | 0.02 |
ZNF469 |
zinc finger protein 469 |
222138 |
0.02 |
| chrX_53117609_53118006 | 0.02 |
TSPYL2 |
TSPY-like 2 |
6244 |
0.19 |
| chr11_11418436_11418587 | 0.02 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
43607 |
0.17 |
| chr8_130957137_130957288 | 0.02 |
FAM49B |
family with sequence similarity 49, member B |
5094 |
0.24 |
| chr21_42217900_42218266 | 0.02 |
DSCAM |
Down syndrome cell adhesion molecule |
982 |
0.71 |
| chr19_47843854_47844005 | 0.02 |
C5AR2 |
complement component 5a receptor 2 |
111 |
0.95 |
| chr6_134323821_134323972 | 0.02 |
TBPL1 |
TBP-like 1 |
49534 |
0.14 |
| chr9_104207627_104207869 | 0.02 |
ALDOB |
aldolase B, fructose-bisphosphate |
9643 |
0.15 |
| chr8_23456525_23456676 | 0.02 |
ENSG00000252067 |
. |
1051 |
0.5 |
| chr10_54463336_54463751 | 0.02 |
RP11-556E13.1 |
|
51626 |
0.16 |
| chr3_30014109_30014328 | 0.02 |
RBMS3-AS1 |
RBMS3 antisense RNA 1 |
38571 |
0.22 |
| chr10_112676510_112676710 | 0.02 |
BBIP1 |
BBSome interacting protein 1 |
2084 |
0.21 |
| chr10_90358006_90358157 | 0.02 |
LIPJ |
lipase, family member J |
3578 |
0.22 |
| chrX_15806325_15806476 | 0.02 |
ZRSR2 |
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
2195 |
0.32 |
| chrX_121981496_121981647 | 0.02 |
ENSG00000212321 |
. |
7292 |
0.34 |
| chr3_149371362_149371801 | 0.02 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
3226 |
0.2 |
| chr5_75764900_75765051 | 0.02 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
64726 |
0.13 |
| chr11_77185235_77185792 | 0.02 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
167 |
0.92 |
| chr12_65032414_65032565 | 0.02 |
RP11-338E21.2 |
|
10365 |
0.12 |
| chr4_188049686_188049901 | 0.02 |
ENSG00000252382 |
. |
271183 |
0.02 |
| chr11_14310125_14310327 | 0.02 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
7179 |
0.24 |
| chr20_30793303_30793454 | 0.02 |
PLAGL2 |
pleiomorphic adenoma gene-like 2 |
2216 |
0.22 |
| chr8_127569289_127570002 | 0.02 |
RP11-89K10.1 |
|
475 |
0.43 |
| chr17_14680546_14680697 | 0.02 |
ENSG00000238806 |
. |
460579 |
0.01 |
| chr3_3843603_3843999 | 0.02 |
LRRN1 |
leucine rich repeat neuronal 1 |
2680 |
0.43 |
| chr21_43739991_43740142 | 0.02 |
TFF3 |
trefoil factor 3 (intestinal) |
4305 |
0.17 |
| chr16_69363414_69364502 | 0.02 |
PDF |
peptide deformylase (mitochondrial) |
540 |
0.59 |
| chr11_75473950_75474101 | 0.02 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
3468 |
0.13 |
| chr17_70358508_70358679 | 0.02 |
SOX9 |
SRY (sex determining region Y)-box 9 |
241432 |
0.02 |
| chr4_154064855_154065006 | 0.02 |
TRIM2 |
tripartite motif containing 2 |
8715 |
0.28 |
| chr12_68661800_68661951 | 0.02 |
IL22 |
interleukin 22 |
14488 |
0.21 |
| chr6_5936596_5936747 | 0.02 |
ENSG00000239472 |
. |
17594 |
0.25 |
| chr9_21563269_21563420 | 0.02 |
MIR31HG |
MIR31 host gene (non-protein coding) |
3676 |
0.23 |
| chr9_3508436_3508948 | 0.02 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
17291 |
0.25 |
| chrX_135258076_135258227 | 0.02 |
FHL1 |
four and a half LIM domains 1 |
6066 |
0.24 |
| chr3_146272331_146272482 | 0.02 |
PLSCR1 |
phospholipid scramblase 1 |
9755 |
0.18 |
| chrX_24043946_24044693 | 0.02 |
KLHL15 |
kelch-like family member 15 |
984 |
0.59 |
| chr7_6481952_6482201 | 0.02 |
DAGLB |
diacylglycerol lipase, beta |
5487 |
0.18 |
| chr9_32741891_32742042 | 0.02 |
ENSG00000221696 |
. |
22328 |
0.18 |
| chr1_198227931_198228157 | 0.02 |
NEK7 |
NIMA-related kinase 7 |
38115 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |