Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
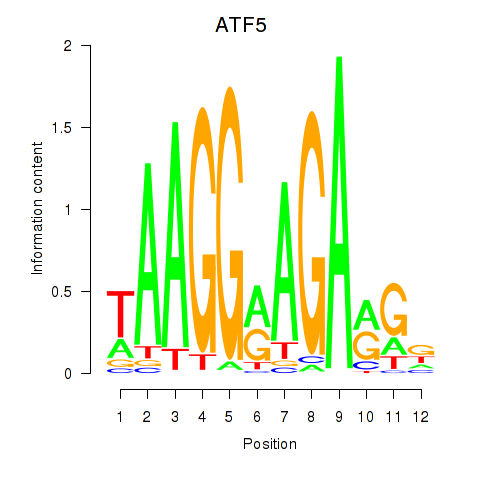
Results for ATF5
Z-value: 0.97
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_50431051_50431205 | ATF5 | 831 | 0.291777 | 0.36 | 3.4e-01 | Click! |
| chr19_50431234_50431474 | ATF5 | 605 | 0.398232 | 0.25 | 5.1e-01 | Click! |
| chr19_50433515_50433677 | ATF5 | 120 | 0.786786 | -0.25 | 5.2e-01 | Click! |
| chr19_50432415_50432712 | ATF5 | 84 | 0.418754 | -0.24 | 5.4e-01 | Click! |
| chr19_50431928_50432406 | ATF5 | 208 | 0.622212 | -0.07 | 8.6e-01 | Click! |
Activity of the ATF5 motif across conditions
Conditions sorted by the z-value of the ATF5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
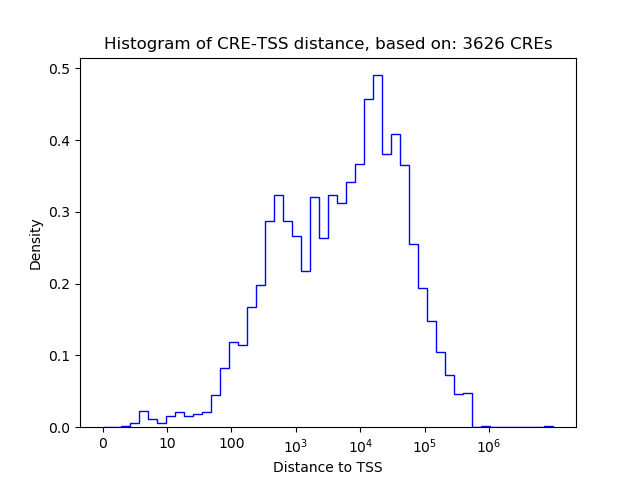
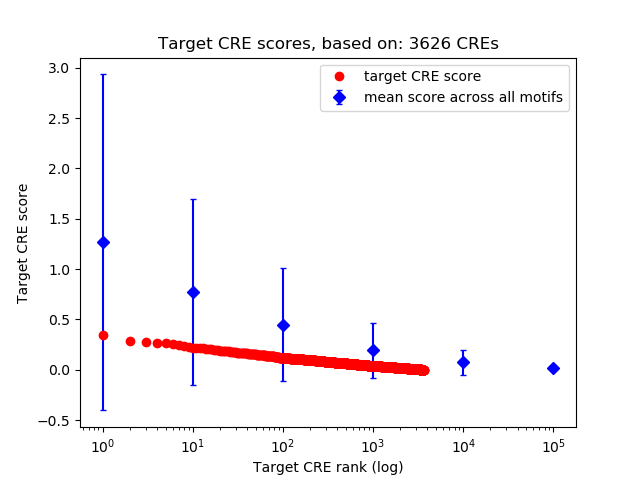
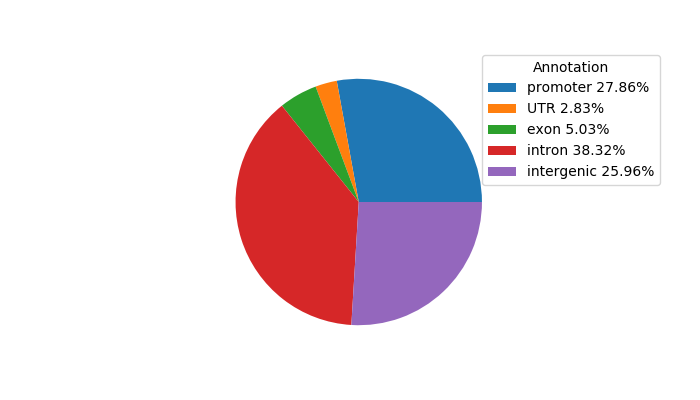
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_82413627_82413778 | 0.35 |
RASGEF1B |
RasGEF domain family, member 1B |
20633 |
0.28 |
| chr1_27890528_27890679 | 0.29 |
RP1-159A19.4 |
|
38287 |
0.11 |
| chr3_194456850_194457149 | 0.27 |
FAM43A |
family with sequence similarity 43, member A |
50377 |
0.11 |
| chr7_115670709_115671033 | 0.27 |
TFEC |
transcription factor EC |
4 |
0.99 |
| chr10_30842846_30843212 | 0.27 |
ENSG00000239744 |
. |
1804 |
0.44 |
| chr2_103506737_103506888 | 0.25 |
TMEM182 |
transmembrane protein 182 |
128340 |
0.06 |
| chr6_5435589_5435798 | 0.25 |
RP1-232P20.1 |
|
22573 |
0.22 |
| chr3_157250255_157250502 | 0.23 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
1017 |
0.61 |
| chr8_91895725_91895978 | 0.22 |
RP11-662G23.1 |
|
24383 |
0.19 |
| chr13_50696260_50696411 | 0.22 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
40028 |
0.15 |
| chr3_171779467_171780048 | 0.22 |
FNDC3B |
fibronectin type III domain containing 3B |
15054 |
0.27 |
| chr13_41576799_41576950 | 0.22 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
16576 |
0.19 |
| chr12_12186755_12186906 | 0.22 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
15948 |
0.26 |
| chr18_56245541_56245920 | 0.21 |
RP11-126O1.2 |
|
21428 |
0.14 |
| chr11_124961540_124961691 | 0.21 |
TMEM218 |
transmembrane protein 218 |
11090 |
0.17 |
| chr1_66258526_66259233 | 0.21 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
15 |
0.99 |
| chr6_84139999_84140706 | 0.20 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
412 |
0.89 |
| chr16_69213517_69213668 | 0.20 |
ENSG00000207083 |
. |
3912 |
0.15 |
| chr10_94517140_94517343 | 0.19 |
ENSG00000201412 |
. |
46876 |
0.14 |
| chr14_103589830_103590609 | 0.19 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
421 |
0.82 |
| chr3_16006218_16006514 | 0.19 |
ENSG00000207815 |
. |
91088 |
0.08 |
| chr2_206750268_206750419 | 0.19 |
ENSG00000201875 |
. |
15011 |
0.24 |
| chr1_197113546_197113697 | 0.19 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
2203 |
0.35 |
| chr5_75601341_75601492 | 0.18 |
RP11-466P24.6 |
|
5871 |
0.31 |
| chr7_26577801_26577952 | 0.18 |
KIAA0087 |
KIAA0087 |
531 |
0.86 |
| chr6_33359684_33360329 | 0.18 |
KIFC1 |
kinesin family member C1 |
368 |
0.77 |
| chr1_245019266_245019417 | 0.18 |
HNRNPU-AS1 |
HNRNPU antisense RNA 1 |
542 |
0.75 |
| chr10_81007808_81008068 | 0.18 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
58037 |
0.14 |
| chr11_57529774_57530544 | 0.17 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
869 |
0.5 |
| chr1_2211042_2211193 | 0.17 |
RP4-713A8.1 |
|
47015 |
0.08 |
| chr11_10627928_10628217 | 0.17 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
13328 |
0.17 |
| chr14_101292536_101293492 | 0.17 |
AL117190.2 |
|
2523 |
0.08 |
| chr1_113585569_113585770 | 0.17 |
LRIG2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
30162 |
0.21 |
| chrX_85249377_85249528 | 0.17 |
ENSG00000199567 |
. |
25160 |
0.22 |
| chr5_172185022_172185186 | 0.17 |
RP11-779O18.1 |
|
125 |
0.95 |
| chr10_6980964_6981115 | 0.17 |
PRKCQ |
protein kinase C, theta |
358776 |
0.01 |
| chr4_55534353_55534504 | 0.17 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
10343 |
0.31 |
| chr6_99426575_99426760 | 0.17 |
FBXL4 |
F-box and leucine-rich repeat protein 4 |
30818 |
0.24 |
| chr2_85815578_85815729 | 0.16 |
VAMP5 |
vesicle-associated membrane protein 5 |
4122 |
0.1 |
| chrX_65220138_65220337 | 0.16 |
RP6-159A1.3 |
|
644 |
0.75 |
| chr7_139268726_139268877 | 0.16 |
CLEC2L |
C-type lectin domain family 2, member L |
59973 |
0.13 |
| chrX_65222772_65222923 | 0.16 |
RP6-159A1.3 |
|
3254 |
0.27 |
| chr9_755961_756198 | 0.16 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
23463 |
0.22 |
| chr1_167618995_167619146 | 0.16 |
RP3-455J7.4 |
|
19159 |
0.18 |
| chr12_95598219_95598370 | 0.16 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
12861 |
0.18 |
| chr4_186951313_186951505 | 0.15 |
ENSG00000222727 |
. |
3669 |
0.24 |
| chr15_52775854_52776075 | 0.15 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
45074 |
0.15 |
| chr21_36280449_36280667 | 0.15 |
RUNX1 |
runt-related transcription factor 1 |
18471 |
0.28 |
| chr5_156706927_156707078 | 0.15 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
5370 |
0.16 |
| chr8_31042072_31042227 | 0.15 |
RP11-363L24.3 |
|
7466 |
0.31 |
| chr2_54724516_54724667 | 0.15 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
40193 |
0.15 |
| chr15_101790171_101790322 | 0.15 |
CHSY1 |
chondroitin sulfate synthase 1 |
1891 |
0.31 |
| chr17_28703666_28703850 | 0.15 |
CPD |
carboxypeptidase D |
2165 |
0.24 |
| chr12_89889035_89889186 | 0.15 |
POC1B |
POC1 centriolar protein B |
1924 |
0.3 |
| chr2_218806529_218806680 | 0.15 |
TNS1 |
tensin 1 |
2189 |
0.35 |
| chr2_236401822_236402641 | 0.15 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
502 |
0.83 |
| chr1_94879075_94879226 | 0.15 |
ABCD3 |
ATP-binding cassette, sub-family D (ALD), member 3 |
4783 |
0.32 |
| chr19_42262167_42262318 | 0.15 |
CEACAM6 |
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
2913 |
0.18 |
| chr4_160100922_160101459 | 0.14 |
ENSG00000264105 |
. |
51144 |
0.13 |
| chr3_112929944_112930573 | 0.14 |
BOC |
BOC cell adhesion associated, oncogene regulated |
71 |
0.98 |
| chr5_32504728_32504940 | 0.14 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
26905 |
0.2 |
| chr16_8725965_8726116 | 0.14 |
METTL22 |
methyltransferase like 22 |
8955 |
0.2 |
| chr3_98609388_98609782 | 0.14 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
10430 |
0.18 |
| chr2_189993459_189993610 | 0.14 |
ENSG00000264725 |
. |
4303 |
0.25 |
| chr15_71148071_71148222 | 0.14 |
LARP6 |
La ribonucleoprotein domain family, member 6 |
1648 |
0.3 |
| chr14_89653839_89653990 | 0.14 |
FOXN3 |
forkhead box N3 |
6826 |
0.3 |
| chr19_32841958_32842109 | 0.14 |
ZNF507 |
zinc finger protein 507 |
5408 |
0.19 |
| chr1_57028471_57028622 | 0.14 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
16695 |
0.27 |
| chr11_128554784_128555228 | 0.14 |
RP11-744N12.3 |
|
1317 |
0.34 |
| chr7_27203057_27203424 | 0.14 |
HOXA9 |
homeobox A9 |
1905 |
0.11 |
| chr3_159483400_159483608 | 0.14 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
612 |
0.7 |
| chr11_118435857_118436109 | 0.14 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
378 |
0.76 |
| chr22_50745196_50745796 | 0.14 |
PLXNB2 |
plexin B2 |
521 |
0.61 |
| chr5_124078870_124079512 | 0.13 |
ZNF608 |
zinc finger protein 608 |
1023 |
0.42 |
| chr4_157893196_157893594 | 0.13 |
PDGFC |
platelet derived growth factor C |
849 |
0.66 |
| chr8_23711375_23711668 | 0.13 |
STC1 |
stanniocalcin 1 |
303 |
0.92 |
| chr2_136776223_136776374 | 0.13 |
AC093391.2 |
|
6263 |
0.25 |
| chr4_175750179_175750687 | 0.13 |
GLRA3 |
glycine receptor, alpha 3 |
32 |
0.99 |
| chr6_138132955_138133132 | 0.13 |
RP11-356I2.4 |
|
13988 |
0.21 |
| chr4_81153527_81153798 | 0.13 |
FGF5 |
fibroblast growth factor 5 |
34091 |
0.16 |
| chr12_45625218_45625410 | 0.13 |
ANO6 |
anoctamin 6 |
15411 |
0.28 |
| chr5_38445912_38446355 | 0.13 |
EGFLAM |
EGF-like, fibronectin type III and laminin G domains |
435 |
0.81 |
| chr2_136631794_136631945 | 0.13 |
MCM6 |
minichromosome maintenance complex component 6 |
2127 |
0.3 |
| chr1_178062269_178062757 | 0.13 |
RASAL2 |
RAS protein activator like 2 |
351 |
0.93 |
| chr5_59837529_59837680 | 0.13 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
53701 |
0.16 |
| chr2_7005603_7006667 | 0.13 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
176 |
0.52 |
| chr3_42066973_42067139 | 0.13 |
ULK4 |
unc-51 like kinase 4 |
63134 |
0.12 |
| chr10_104536560_104536809 | 0.13 |
WBP1L |
WW domain binding protein 1-like |
678 |
0.63 |
| chr1_98496294_98496445 | 0.12 |
ENSG00000225206 |
. |
14538 |
0.29 |
| chr12_91499433_91499620 | 0.12 |
LUM |
lumican |
6082 |
0.24 |
| chr8_95563818_95563969 | 0.12 |
KIAA1429 |
KIAA1429 |
1795 |
0.3 |
| chr20_57678404_57678555 | 0.12 |
SLMO2 |
slowmo homolog 2 (Drosophila) |
60515 |
0.1 |
| chr12_54652241_54652451 | 0.12 |
CBX5 |
chromobox homolog 5 |
135 |
0.9 |
| chr6_33808044_33808280 | 0.12 |
MLN |
motilin |
36374 |
0.15 |
| chr3_119349777_119349928 | 0.12 |
PLA1A |
phospholipase A1 member A |
22108 |
0.13 |
| chr7_92258314_92258465 | 0.12 |
FAM133B |
family with sequence similarity 133, member B |
38681 |
0.16 |
| chr12_45445409_45445593 | 0.12 |
DBX2 |
developing brain homeobox 2 |
619 |
0.54 |
| chr6_105402236_105402387 | 0.12 |
LIN28B |
lin-28 homolog B (C. elegans) |
2612 |
0.35 |
| chr21_36263531_36263991 | 0.12 |
RUNX1 |
runt-related transcription factor 1 |
1674 |
0.53 |
| chr15_64671727_64672232 | 0.12 |
KIAA0101 |
KIAA0101 |
1658 |
0.23 |
| chr3_106568854_106569005 | 0.12 |
ENSG00000252626 |
. |
157486 |
0.04 |
| chr8_8421576_8421727 | 0.12 |
ENSG00000263616 |
. |
18514 |
0.24 |
| chr1_52519790_52519941 | 0.12 |
TXNDC12 |
thioredoxin domain containing 12 (endoplasmic reticulum) |
998 |
0.36 |
| chr10_92644478_92644629 | 0.12 |
ENSG00000222305 |
. |
4617 |
0.19 |
| chr4_72159796_72159986 | 0.12 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
44879 |
0.2 |
| chr10_34149976_34150160 | 0.12 |
ENSG00000199200 |
. |
340934 |
0.01 |
| chr4_71940368_71940554 | 0.12 |
DCK |
deoxycytidine kinase |
81106 |
0.1 |
| chr17_48504516_48504676 | 0.12 |
ACSF2 |
acyl-CoA synthetase family member 2 |
983 |
0.39 |
| chr7_116504981_116505132 | 0.12 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
2402 |
0.29 |
| chr3_148583159_148583310 | 0.12 |
CPA3 |
carboxypeptidase A3 (mast cell) |
191 |
0.96 |
| chr1_26158474_26158625 | 0.12 |
RP1-317E23.7 |
|
76 |
0.94 |
| chr17_72869969_72870224 | 0.12 |
FDXR |
ferredoxin reductase |
940 |
0.41 |
| chr5_52542179_52542330 | 0.11 |
MOCS2 |
molybdenum cofactor synthesis 2 |
136361 |
0.05 |
| chr9_13278507_13279169 | 0.11 |
RP11-272P10.2 |
|
348 |
0.67 |
| chr8_63160920_63161162 | 0.11 |
NKAIN3 |
Na+/K+ transporting ATPase interacting 3 |
460 |
0.91 |
| chr3_159562165_159562360 | 0.11 |
SCHIP1 |
schwannomin interacting protein 1 |
4612 |
0.21 |
| chr6_121895181_121895332 | 0.11 |
ENSG00000222659 |
. |
6411 |
0.25 |
| chr4_91239201_91239404 | 0.11 |
ENSG00000252087 |
. |
51972 |
0.17 |
| chr13_24006860_24007105 | 0.11 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
859 |
0.74 |
| chr10_128258751_128258911 | 0.11 |
ENSG00000221717 |
. |
13198 |
0.26 |
| chr8_6603359_6603510 | 0.11 |
ENSG00000266038 |
. |
749 |
0.69 |
| chr12_93576211_93576362 | 0.11 |
RP11-511B23.2 |
|
33164 |
0.17 |
| chr1_110933283_110933653 | 0.11 |
SLC16A4 |
solute carrier family 16, member 4 |
180 |
0.92 |
| chr9_212362_212513 | 0.11 |
DOCK8 |
dedicator of cytokinesis 8 |
2428 |
0.26 |
| chr11_33283703_33283854 | 0.11 |
HIPK3 |
homeodomain interacting protein kinase 3 |
3901 |
0.27 |
| chr4_185292014_185292165 | 0.11 |
IRF2 |
interferon regulatory factor 2 |
47655 |
0.14 |
| chr3_160203157_160203402 | 0.11 |
TRIM59 |
tripartite motif containing 59 |
282 |
0.89 |
| chr16_73069914_73070167 | 0.11 |
ZFHX3 |
zinc finger homeobox 3 |
12234 |
0.21 |
| chr5_148185821_148186248 | 0.11 |
ADRB2 |
adrenoceptor beta 2, surface |
20122 |
0.24 |
| chr14_24805236_24805387 | 0.11 |
ADCY4 |
adenylate cyclase 4 |
1034 |
0.21 |
| chr5_371927_372078 | 0.11 |
AHRR |
aryl-hydrocarbon receptor repressor |
28359 |
0.15 |
| chr2_43232535_43232725 | 0.11 |
ENSG00000207087 |
. |
86002 |
0.1 |
| chr6_51928260_51928411 | 0.11 |
ENSG00000264206 |
. |
1831 |
0.37 |
| chrX_65896718_65896962 | 0.11 |
ENSG00000222667 |
. |
634 |
0.81 |
| chr16_48647179_48647330 | 0.11 |
N4BP1 |
NEDD4 binding protein 1 |
3134 |
0.25 |
| chr17_55063311_55063462 | 0.11 |
RP5-1107A17.4 |
|
3276 |
0.17 |
| chr2_145278154_145278598 | 0.11 |
ZEB2-AS1 |
ZEB2 antisense RNA 1 |
203 |
0.55 |
| chr8_106536977_106537128 | 0.11 |
ZFPM2 |
zinc finger protein, FOG family member 2 |
4060 |
0.36 |
| chr17_76907300_76907451 | 0.11 |
CTD-2373H9.5 |
|
8030 |
0.14 |
| chr19_41913570_41913729 | 0.11 |
BCKDHA |
branched chain keto acid dehydrogenase E1, alpha polypeptide |
2978 |
0.12 |
| chr7_30199849_30200000 | 0.11 |
AC007036.5 |
|
2777 |
0.24 |
| chr11_105481715_105481968 | 0.11 |
GRIA4 |
glutamate receptor, ionotropic, AMPA 4 |
116 |
0.98 |
| chr7_18376735_18376886 | 0.11 |
ENSG00000223030 |
. |
39848 |
0.17 |
| chr1_78355822_78355973 | 0.11 |
NEXN-AS1 |
NEXN antisense RNA 1 |
673 |
0.62 |
| chr12_124475208_124475359 | 0.11 |
ZNF664 |
zinc finger protein 664 |
17441 |
0.08 |
| chr13_113704257_113704408 | 0.11 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
5167 |
0.16 |
| chr1_151286527_151286678 | 0.11 |
PI4KB |
phosphatidylinositol 4-kinase, catalytic, beta |
12122 |
0.09 |
| chr2_212025965_212026310 | 0.11 |
ENSG00000199585 |
. |
374241 |
0.01 |
| chr22_38438659_38438835 | 0.11 |
RP5-1039K5.16 |
|
8512 |
0.11 |
| chr1_202474380_202474681 | 0.11 |
ENSG00000253042 |
. |
22025 |
0.18 |
| chr8_38585652_38586394 | 0.11 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
82 |
0.97 |
| chr8_16998847_16999036 | 0.11 |
ZDHHC2 |
zinc finger, DHHC-type containing 2 |
14597 |
0.24 |
| chr1_115397478_115398199 | 0.11 |
SYCP1 |
synaptonemal complex protein 1 |
84 |
0.98 |
| chr8_146204034_146204185 | 0.10 |
RP5-1047A19.4 |
|
14191 |
0.15 |
| chr1_114525510_114525669 | 0.10 |
OLFML3 |
olfactomedin-like 3 |
2894 |
0.24 |
| chr5_124079655_124079839 | 0.10 |
ZNF608 |
zinc finger protein 608 |
467 |
0.74 |
| chr7_133039894_133040045 | 0.10 |
ENSG00000238844 |
. |
92484 |
0.09 |
| chr11_26305888_26306039 | 0.10 |
ANO3 |
anoctamin 3 |
25183 |
0.28 |
| chr1_167518297_167518545 | 0.10 |
CREG1 |
cellular repressor of E1A-stimulated genes 1 |
4583 |
0.23 |
| chr3_127453865_127454048 | 0.10 |
MGLL |
monoglyceride lipase |
1244 |
0.53 |
| chr2_27434850_27435940 | 0.10 |
SLC5A6 |
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
5 |
0.66 |
| chr17_31255277_31255844 | 0.10 |
TMEM98 |
transmembrane protein 98 |
332 |
0.89 |
| chr7_27191312_27191528 | 0.10 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
1749 |
0.13 |
| chr14_55239682_55239899 | 0.10 |
SAMD4A |
sterile alpha motif domain containing 4A |
18239 |
0.24 |
| chr16_22200871_22201892 | 0.10 |
EEF2K |
eukaryotic elongation factor-2 kinase |
16222 |
0.16 |
| chr9_111578370_111578673 | 0.10 |
ACTL7B |
actin-like 7B |
40718 |
0.16 |
| chr14_103316071_103316226 | 0.10 |
ENSG00000206969 |
. |
16045 |
0.15 |
| chr22_39601535_39601686 | 0.10 |
PDGFB |
platelet-derived growth factor beta polypeptide |
35304 |
0.11 |
| chr7_12251046_12251631 | 0.10 |
TMEM106B |
transmembrane protein 106B |
389 |
0.93 |
| chr4_156680188_156681171 | 0.10 |
GUCY1B3 |
guanylate cyclase 1, soluble, beta 3 |
68 |
0.98 |
| chr20_43729394_43729952 | 0.10 |
KCNS1 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
77 |
0.96 |
| chr16_30770390_30770590 | 0.10 |
C16orf93 |
chromosome 16 open reading frame 93 |
555 |
0.55 |
| chr15_75416074_75416396 | 0.10 |
ENSG00000252722 |
. |
2228 |
0.28 |
| chrX_149596043_149596194 | 0.10 |
MAMLD1 |
mastermind-like domain containing 1 |
6845 |
0.28 |
| chr8_11415915_11416066 | 0.10 |
RP11-148O21.4 |
|
459 |
0.72 |
| chr3_37114235_37114598 | 0.10 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
10825 |
0.17 |
| chr1_101727926_101728077 | 0.10 |
RP4-575N6.5 |
|
19287 |
0.16 |
| chr11_72999273_72999424 | 0.10 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
1104 |
0.43 |
| chr7_106377259_106377410 | 0.10 |
CTB-111H14.1 |
|
4439 |
0.28 |
| chr21_43884438_43884684 | 0.10 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
31567 |
0.1 |
| chr13_32518987_32519138 | 0.10 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
7655 |
0.28 |
| chr20_2236731_2236882 | 0.10 |
TGM3 |
transglutaminase 3 |
39841 |
0.17 |
| chr5_59817644_59817795 | 0.10 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
33816 |
0.22 |
| chr12_26379869_26380400 | 0.10 |
SSPN |
sarcospan |
31528 |
0.16 |
| chr2_64878829_64879050 | 0.10 |
SERTAD2 |
SERTA domain containing 2 |
2108 |
0.38 |
| chr9_71510539_71510690 | 0.10 |
ENSG00000207000 |
. |
40281 |
0.16 |
| chr1_46017329_46017598 | 0.10 |
AKR1A1 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
723 |
0.6 |
| chr17_40934580_40934731 | 0.10 |
WNK4 |
WNK lysine deficient protein kinase 4 |
1959 |
0.15 |
| chr16_23869995_23870146 | 0.10 |
PRKCB |
protein kinase C, beta |
21526 |
0.22 |
| chr5_180590682_180590909 | 0.10 |
OR2V2 |
olfactory receptor, family 2, subfamily V, member 2 |
8852 |
0.1 |
| chr2_54649975_54650126 | 0.10 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
33372 |
0.19 |
| chr17_27462701_27462852 | 0.10 |
MYO18A |
myosin XVIIIA |
4660 |
0.17 |
| chr8_63998951_63999120 | 0.10 |
TTPA |
tocopherol (alpha) transfer protein |
423 |
0.86 |
| chr7_120646497_120646648 | 0.10 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
16896 |
0.19 |
| chr12_112860746_112860897 | 0.10 |
PTPN11 |
protein tyrosine phosphatase, non-receptor type 11 |
4103 |
0.22 |
| chr17_79317268_79317735 | 0.10 |
TMEM105 |
transmembrane protein 105 |
13027 |
0.13 |
| chr18_56246037_56246225 | 0.10 |
ENSG00000252284 |
. |
21732 |
0.14 |
| chr20_50527044_50527195 | 0.10 |
ENSG00000252654 |
. |
5611 |
0.21 |
| chr5_95005032_95005183 | 0.10 |
SPATA9 |
spermatogenesis associated 9 |
13553 |
0.16 |
| chr1_156868658_156868809 | 0.10 |
PEAR1 |
platelet endothelial aggregation receptor 1 |
5210 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |