Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
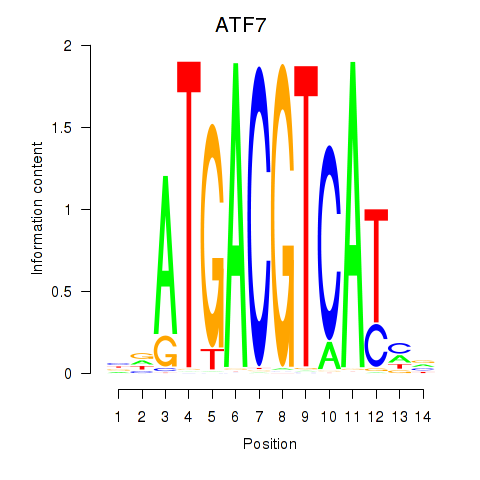
Results for ATF7
Z-value: 0.82
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | activating transcription factor 7 |
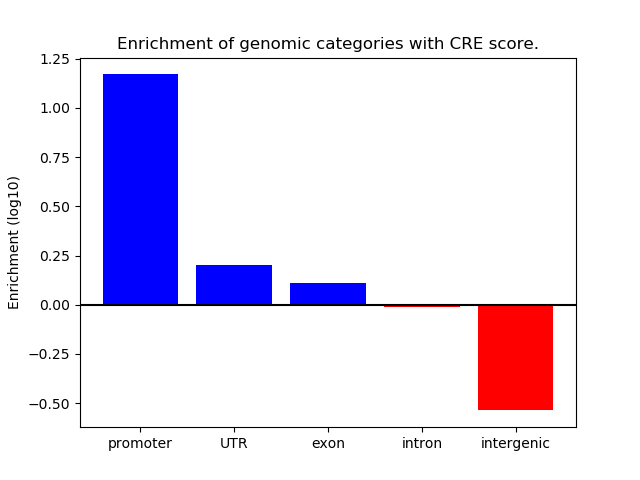
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_53954959_53955110 | ATF7 | 39771 | 0.088898 | -0.66 | 5.5e-02 | Click! |
| chr12_53985363_53985546 | ATF7 | 9351 | 0.153411 | -0.62 | 7.4e-02 | Click! |
| chr12_54019958_54020201 | ATF7 | 68 | 0.797129 | 0.62 | 7.4e-02 | Click! |
| chr12_54020323_54020579 | ATF7 | 252 | 0.783018 | 0.59 | 9.7e-02 | Click! |
| chr12_53985687_53985890 | ATF7 | 9017 | 0.154243 | -0.55 | 1.2e-01 | Click! |
Activity of the ATF7 motif across conditions
Conditions sorted by the z-value of the ATF7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
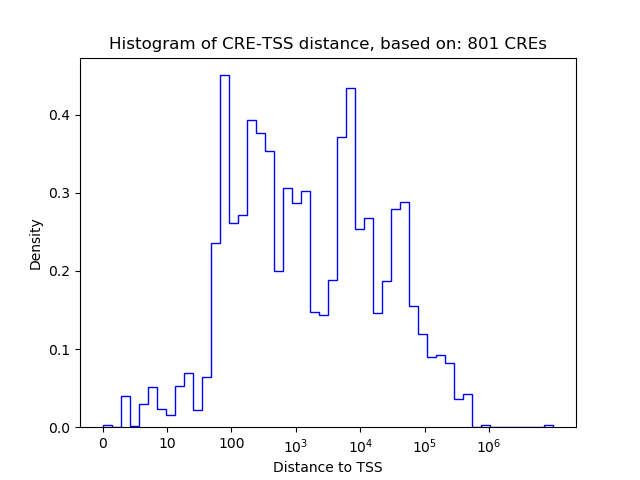
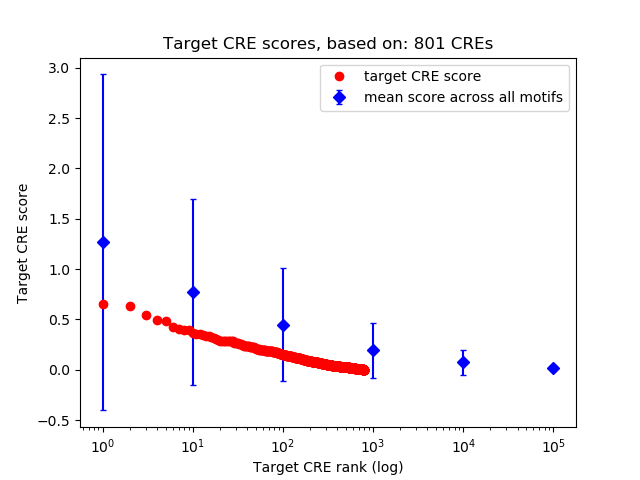
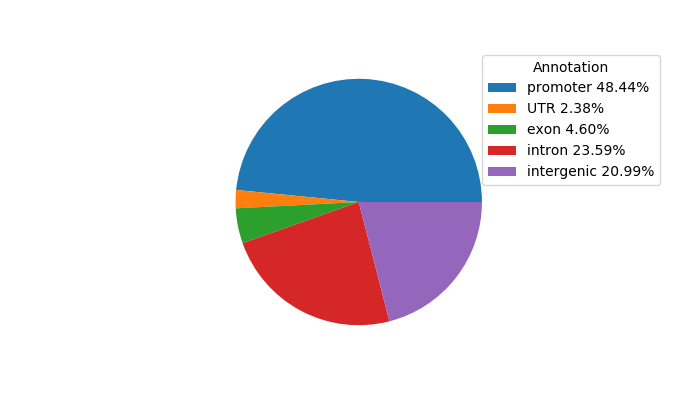
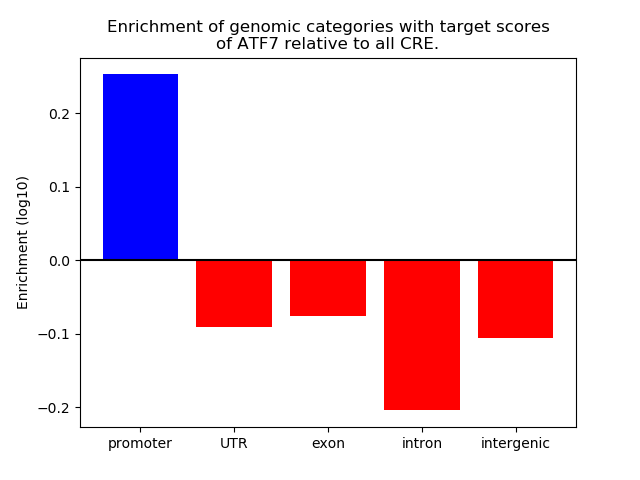
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_21967077_21967500 | 0.65 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
356 |
0.79 |
| chr5_176881613_176881764 | 0.63 |
PRR7 |
proline rich 7 (synaptic) |
6353 |
0.1 |
| chr2_28582064_28582304 | 0.55 |
FOSL2 |
FOS-like antigen 2 |
33485 |
0.14 |
| chr19_51014778_51015051 | 0.50 |
JOSD2 |
Josephin domain containing 2 |
304 |
0.8 |
| chr20_16926810_16926961 | 0.48 |
ENSG00000212165 |
. |
36793 |
0.22 |
| chr16_31142045_31142196 | 0.42 |
RP11-388M20.2 |
|
634 |
0.42 |
| chr17_4046800_4047053 | 0.40 |
CYB5D2 |
cytochrome b5 domain containing 2 |
70 |
0.9 |
| chr12_56521312_56521715 | 0.39 |
ESYT1 |
extended synaptotagmin-like protein 1 |
327 |
0.64 |
| chr19_17666323_17666474 | 0.39 |
COLGALT1 |
collagen beta(1-O)galactosyltransferase 1 |
5 |
0.96 |
| chr19_8454704_8454922 | 0.36 |
RAB11B |
RAB11B, member RAS oncogene family |
52 |
0.65 |
| chr20_19915434_19915955 | 0.36 |
RIN2 |
Ras and Rab interactor 2 |
45484 |
0.16 |
| chr4_15721295_15721446 | 0.35 |
BST1 |
bone marrow stromal cell antigen 1 |
4419 |
0.23 |
| chr6_91006489_91006667 | 0.35 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
49 |
0.98 |
| chr5_73730259_73730491 | 0.33 |
ENSG00000244326 |
. |
116564 |
0.06 |
| chr20_34203605_34204165 | 0.33 |
SPAG4 |
sperm associated antigen 4 |
71 |
0.95 |
| chr9_95895594_95896005 | 0.32 |
NINJ1 |
ninjurin 1 |
771 |
0.63 |
| chr9_137347969_137348661 | 0.32 |
RXRA |
retinoid X receptor, alpha |
49887 |
0.15 |
| chr2_74781498_74781793 | 0.30 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
213 |
0.73 |
| chr16_86817661_86817812 | 0.30 |
FOXL1 |
forkhead box L1 |
205621 |
0.02 |
| chr19_42900892_42901243 | 0.29 |
LIPE-AS1 |
LIPE antisense RNA 1 |
213 |
0.88 |
| chr19_46009815_46010009 | 0.29 |
VASP |
vasodilator-stimulated phosphoprotein |
75 |
0.95 |
| chr11_18720267_18720541 | 0.29 |
TMEM86A |
transmembrane protein 86A |
66 |
0.96 |
| chr14_103988458_103988873 | 0.29 |
CKB |
creatine kinase, brain |
368 |
0.73 |
| chr21_46898660_46898949 | 0.28 |
COL18A1 |
collagen, type XVIII, alpha 1 |
11385 |
0.19 |
| chr10_99078775_99079092 | 0.28 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
89 |
0.95 |
| chr1_228353838_228353989 | 0.28 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
397 |
0.6 |
| chr17_18528902_18529084 | 0.28 |
CCDC144B |
coiled-coil domain containing 144B (pseudogene) |
72 |
0.96 |
| chr2_8144573_8144775 | 0.28 |
ENSG00000221255 |
. |
427702 |
0.01 |
| chr10_105728102_105728505 | 0.27 |
SLK |
STE20-like kinase |
1344 |
0.4 |
| chr3_170075366_170075517 | 0.27 |
SKIL |
SKI-like oncogene |
62 |
0.98 |
| chr1_228356433_228356584 | 0.27 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
2992 |
0.14 |
| chr7_38217813_38218059 | 0.26 |
STARD3NL |
STARD3 N-terminal like |
17 |
0.99 |
| chr18_2654684_2655529 | 0.26 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
631 |
0.67 |
| chr8_145555468_145555619 | 0.25 |
SCRT1 |
scratch family zinc finger 1 |
4400 |
0.08 |
| chr22_42228451_42228869 | 0.25 |
SREBF2 |
sterol regulatory element binding transcription factor 2 |
449 |
0.71 |
| chr9_35617185_35617463 | 0.24 |
CD72 |
CD72 molecule |
1043 |
0.3 |
| chr11_110168760_110168911 | 0.24 |
RDX |
radixin |
1388 |
0.59 |
| chr5_170739718_170739894 | 0.24 |
TLX3 |
T-cell leukemia homeobox 3 |
3518 |
0.22 |
| chr4_159131007_159131158 | 0.24 |
TMEM144 |
transmembrane protein 144 |
264 |
0.91 |
| chr19_55919228_55919426 | 0.24 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
182 |
0.86 |
| chr16_85045097_85045598 | 0.24 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
206 |
0.95 |
| chr22_51059513_51059819 | 0.23 |
ARSA |
arylsulfatase A |
6917 |
0.1 |
| chr15_59658629_59658780 | 0.23 |
FAM81A |
family with sequence similarity 81, member A |
6188 |
0.13 |
| chr8_33422545_33422696 | 0.23 |
RNF122 |
ring finger protein 122 |
2023 |
0.25 |
| chr16_88859439_88859692 | 0.22 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
7946 |
0.09 |
| chr5_107005011_107005570 | 0.22 |
EFNA5 |
ephrin-A5 |
1038 |
0.67 |
| chr11_82782830_82782981 | 0.22 |
RAB30 |
RAB30, member RAS oncogene family |
3 |
0.98 |
| chr11_108535331_108535645 | 0.22 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
264 |
0.95 |
| chr16_31471330_31471687 | 0.22 |
ARMC5 |
armadillo repeat containing 5 |
226 |
0.67 |
| chr2_43822335_43823890 | 0.21 |
THADA |
thyroid adenoma associated |
1 |
0.98 |
| chr20_12990547_12990774 | 0.21 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
663 |
0.83 |
| chr17_6926858_6927139 | 0.20 |
BCL6B |
B-cell CLL/lymphoma 6, member B |
202 |
0.83 |
| chr6_36524849_36525000 | 0.20 |
STK38 |
serine/threonine kinase 38 |
9677 |
0.14 |
| chr14_69260124_69260503 | 0.20 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
356 |
0.9 |
| chr21_45209367_45209684 | 0.20 |
RRP1 |
ribosomal RNA processing 1 |
131 |
0.95 |
| chr22_36519910_36520146 | 0.20 |
APOL3 |
apolipoprotein L, 3 |
36803 |
0.17 |
| chr3_182512816_182512967 | 0.20 |
ATP11B |
ATPase, class VI, type 11B |
1600 |
0.36 |
| chr11_33183160_33183580 | 0.20 |
CSTF3-AS1 |
CSTF3 antisense RNA 1 (head to head) |
136 |
0.7 |
| chr12_71833497_71833776 | 0.20 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
86 |
0.97 |
| chr7_4764745_4765032 | 0.20 |
FOXK1 |
forkhead box K1 |
15602 |
0.18 |
| chr2_108994212_108994393 | 0.20 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
166 |
0.96 |
| chr17_2496882_2497033 | 0.20 |
PAFAH1B1 |
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
83 |
0.97 |
| chr1_17630726_17630943 | 0.19 |
PADI4 |
peptidyl arginine deiminase, type IV |
3856 |
0.2 |
| chr14_55738060_55738282 | 0.19 |
FBXO34 |
F-box protein 34 |
150 |
0.97 |
| chr9_130860266_130860759 | 0.19 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
71 |
0.93 |
| chr15_89905376_89905703 | 0.19 |
ENSG00000207819 |
. |
5709 |
0.19 |
| chr19_45349520_45349839 | 0.19 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
104 |
0.94 |
| chr9_139117671_139117822 | 0.19 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
1023 |
0.52 |
| chr12_120729731_120730139 | 0.19 |
ENSG00000202538 |
. |
229 |
0.83 |
| chr7_27232397_27232921 | 0.19 |
HOXA11-AS |
HOXA11 antisense RNA |
5221 |
0.07 |
| chr20_15773685_15773836 | 0.18 |
MACROD2 |
MACRO domain containing 2 |
69690 |
0.14 |
| chr22_35805907_35806058 | 0.18 |
MCM5 |
minichromosome maintenance complex component 5 |
9558 |
0.2 |
| chr6_159519573_159519904 | 0.18 |
TAGAP |
T-cell activation RhoGTPase activating protein |
53554 |
0.11 |
| chr1_234176768_234176919 | 0.18 |
RP11-488L4.1 |
|
89990 |
0.09 |
| chr9_34665613_34666108 | 0.18 |
RP11-195F19.5 |
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
165 |
0.52 |
| chr1_155292308_155292690 | 0.18 |
RUSC1-AS1 |
RUSC1 antisense RNA 1 |
1164 |
0.22 |
| chr17_7165860_7166195 | 0.18 |
CLDN7 |
claudin 7 |
230 |
0.78 |
| chr8_39043803_39043954 | 0.18 |
ADAM32 |
ADAM metallopeptidase domain 32 |
78403 |
0.1 |
| chr10_106089117_106089268 | 0.18 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
510 |
0.71 |
| chr1_38465984_38466787 | 0.18 |
FHL3 |
four and a half LIM domains 3 |
4792 |
0.14 |
| chr5_112849172_112849403 | 0.17 |
YTHDC2 |
YTH domain containing 2 |
93 |
0.97 |
| chr6_3157575_3157860 | 0.17 |
TUBB2A |
tubulin, beta 2A class IIa |
43 |
0.97 |
| chr16_70730957_70731108 | 0.17 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
1536 |
0.32 |
| chr21_44597084_44597458 | 0.17 |
CRYAA |
crystallin, alpha A |
7005 |
0.17 |
| chr3_120277883_120278034 | 0.17 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
37198 |
0.19 |
| chr9_94710782_94711296 | 0.17 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
123 |
0.98 |
| chr8_13129460_13129611 | 0.17 |
DLC1 |
deleted in liver cancer 1 |
4520 |
0.3 |
| chr2_138880085_138880236 | 0.17 |
HNMT |
histamine N-methyltransferase |
158117 |
0.04 |
| chr12_54378782_54378933 | 0.17 |
HOXC10 |
homeobox C10 |
8 |
0.71 |
| chr3_29228940_29229091 | 0.17 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
93458 |
0.09 |
| chr19_1021594_1021873 | 0.16 |
ENSG00000207357 |
. |
212 |
0.73 |
| chr3_33759550_33759763 | 0.16 |
CLASP2 |
cytoplasmic linker associated protein 2 |
115 |
0.98 |
| chrX_54665940_54666267 | 0.16 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
109428 |
0.06 |
| chr2_238609518_238609709 | 0.16 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
8631 |
0.24 |
| chr16_87896068_87896219 | 0.16 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
1102 |
0.49 |
| chr7_100209432_100209679 | 0.16 |
MOSPD3 |
motile sperm domain containing 3 |
170 |
0.88 |
| chr16_49737112_49737263 | 0.16 |
ENSG00000221134 |
. |
24056 |
0.21 |
| chr16_12086577_12086728 | 0.15 |
RP11-166B2.7 |
|
14945 |
0.11 |
| chrX_98854100_98854251 | 0.15 |
ENSG00000252430 |
. |
213901 |
0.03 |
| chr2_64957643_64958616 | 0.15 |
ENSG00000253082 |
. |
45109 |
0.14 |
| chr17_36740988_36741255 | 0.15 |
CTB-58E17.2 |
|
16181 |
0.13 |
| chr19_39175153_39175304 | 0.15 |
CTB-186G2.4 |
|
9018 |
0.12 |
| chr6_44355057_44355355 | 0.15 |
CDC5L |
cell division cycle 5-like |
56 |
0.97 |
| chr1_169863342_169863522 | 0.15 |
SCYL3 |
SCY1-like 3 (S. cerevisiae) |
24 |
0.98 |
| chr19_2956542_2956693 | 0.15 |
ZNF77 |
zinc finger protein 77 |
11686 |
0.12 |
| chr7_140396423_140396944 | 0.15 |
NDUFB2 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
105 |
0.63 |
| chr12_7282858_7283009 | 0.14 |
RP11-273B20.1 |
|
85 |
0.56 |
| chr18_74205501_74206173 | 0.14 |
ZNF516 |
zinc finger protein 516 |
1309 |
0.32 |
| chr1_212963145_212963296 | 0.14 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
1703 |
0.3 |
| chr20_3782117_3782268 | 0.14 |
CDC25B |
cell division cycle 25B |
5114 |
0.13 |
| chr4_170580994_170582031 | 0.14 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
299 |
0.93 |
| chr19_55574631_55574782 | 0.14 |
RDH13 |
retinol dehydrogenase 13 (all-trans/9-cis) |
162 |
0.91 |
| chr20_56293559_56293931 | 0.14 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
7204 |
0.28 |
| chr8_135844464_135844657 | 0.14 |
ENSG00000199153 |
. |
27372 |
0.21 |
| chr1_29213318_29213476 | 0.14 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
206 |
0.93 |
| chr1_145396692_145396864 | 0.14 |
ENSG00000201558 |
. |
14021 |
0.13 |
| chr1_22379888_22380679 | 0.14 |
CDC42 |
cell division cycle 42 |
493 |
0.71 |
| chr3_101405604_101405866 | 0.14 |
RPL24 |
ribosomal protein L24 |
109 |
0.94 |
| chr11_118122819_118123419 | 0.14 |
MPZL3 |
myelin protein zero-like 3 |
54 |
0.96 |
| chr5_130971941_130972134 | 0.14 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
1108 |
0.65 |
| chr12_27939182_27939333 | 0.14 |
RP11-860B13.1 |
|
5257 |
0.13 |
| chr1_181056884_181057035 | 0.13 |
IER5 |
immediate early response 5 |
679 |
0.74 |
| chr3_184053679_184053952 | 0.13 |
FAM131A |
family with sequence similarity 131, member A |
82 |
0.94 |
| chr6_27832819_27832970 | 0.13 |
HIST1H2AL |
histone cluster 1, H2al |
140 |
0.87 |
| chr20_34203389_34203540 | 0.13 |
SPAG4 |
sperm associated antigen 4 |
350 |
0.8 |
| chr17_79093601_79093928 | 0.13 |
ENSG00000207736 |
. |
5409 |
0.1 |
| chr7_96634662_96635012 | 0.13 |
DLX6 |
distal-less homeobox 6 |
23 |
0.95 |
| chr8_142275255_142276084 | 0.13 |
RP11-10J21.3 |
Uncharacterized protein |
11005 |
0.14 |
| chr8_69155638_69155789 | 0.13 |
ENSG00000252210 |
. |
20076 |
0.25 |
| chr19_6393244_6393475 | 0.13 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
112 |
0.91 |
| chr3_31577931_31578214 | 0.13 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
3790 |
0.36 |
| chr14_74352825_74353319 | 0.13 |
ZNF410 |
zinc finger protein 410 |
248 |
0.86 |
| chr19_10974082_10974233 | 0.13 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
8032 |
0.12 |
| chr4_77875208_77875359 | 0.13 |
SEPT11 |
septin 11 |
4268 |
0.27 |
| chr8_145561219_145561461 | 0.13 |
SCRT1 |
scratch family zinc finger 1 |
1397 |
0.19 |
| chr6_28864112_28865009 | 0.12 |
HCG14 |
HLA complex group 14 (non-protein coding) |
253 |
0.89 |
| chr14_63749678_63749829 | 0.12 |
ENSG00000265711 |
. |
50223 |
0.15 |
| chr2_47403720_47403910 | 0.12 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
75 |
0.98 |
| chr8_142180071_142180222 | 0.12 |
DENND3 |
DENN/MADD domain containing 3 |
8074 |
0.2 |
| chr8_74792637_74792788 | 0.12 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
1567 |
0.43 |
| chr7_151696730_151696881 | 0.12 |
GALNT11 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
25973 |
0.14 |
| chr19_13056278_13056490 | 0.12 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
285 |
0.69 |
| chr17_58603607_58603878 | 0.12 |
RP11-15E18.1 |
|
82 |
0.61 |
| chr21_16665827_16665978 | 0.12 |
NRIP1 |
nuclear receptor interacting protein 1 |
228581 |
0.02 |
| chr2_78182087_78182238 | 0.12 |
LRRTM4 |
leucine rich repeat transmembrane neuronal 4 |
361840 |
0.01 |
| chr21_43619409_43619560 | 0.12 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
315 |
0.9 |
| chr11_13238316_13238540 | 0.12 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
59771 |
0.15 |
| chr13_110383367_110383518 | 0.12 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
2813 |
0.38 |
| chr10_63779003_63779237 | 0.11 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
29850 |
0.22 |
| chr16_68056878_68057148 | 0.11 |
DUS2 |
dihydrouridine synthase 2 |
143 |
0.85 |
| chr1_153917120_153917271 | 0.11 |
DENND4B |
DENN/MADD domain containing 4B |
550 |
0.58 |
| chr8_41625594_41625745 | 0.11 |
ANK1 |
ankyrin 1, erythrocytic |
29471 |
0.15 |
| chr1_28655191_28655470 | 0.11 |
MED18 |
mediator complex subunit 18 |
219 |
0.92 |
| chr12_123201117_123201268 | 0.11 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
247 |
0.89 |
| chr11_77113152_77113303 | 0.11 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
9722 |
0.24 |
| chr13_32889728_32890299 | 0.11 |
BRCA2 |
breast cancer 2, early onset |
371 |
0.59 |
| chr12_25538523_25539010 | 0.11 |
ENSG00000201439 |
. |
18596 |
0.25 |
| chr22_39745318_39745552 | 0.11 |
SYNGR1 |
synaptogyrin 1 |
495 |
0.71 |
| chr2_41495595_41495746 | 0.11 |
ENSG00000221372 |
. |
327856 |
0.01 |
| chr11_1404301_1404651 | 0.11 |
BRSK2 |
BR serine/threonine kinase 2 |
6653 |
0.2 |
| chr18_21573835_21574026 | 0.11 |
TTC39C |
tetratricopeptide repeat domain 39C |
1193 |
0.47 |
| chr17_75197356_75197507 | 0.11 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
5466 |
0.24 |
| chr14_100112036_100112187 | 0.11 |
HHIPL1 |
HHIP-like 1 |
631 |
0.72 |
| chrX_152337908_152338131 | 0.11 |
PNMA6A |
paraneoplastic Ma antigen family member 6A |
282 |
0.91 |
| chr22_26875322_26876099 | 0.11 |
HPS4 |
Hermansky-Pudlak syndrome 4 |
63 |
0.96 |
| chr16_2563812_2564354 | 0.10 |
ATP6V0C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
106 |
0.49 |
| chr1_167569579_167569730 | 0.10 |
RCSD1 |
RCSD domain containing 1 |
29676 |
0.15 |
| chr17_4643181_4643351 | 0.10 |
CXCL16 |
chemokine (C-X-C motif) ligand 16 |
49 |
0.49 |
| chr1_91183266_91183570 | 0.10 |
BARHL2 |
BarH-like homeobox 2 |
624 |
0.83 |
| chr3_122513577_122513821 | 0.10 |
DIRC2 |
disrupted in renal carcinoma 2 |
57 |
0.95 |
| chr16_27378896_27379134 | 0.10 |
IL4R |
interleukin 4 receptor |
15019 |
0.19 |
| chrX_57931578_57932092 | 0.10 |
ZXDA |
zinc finger, X-linked, duplicated A |
5232 |
0.36 |
| chr1_161495345_161495496 | 0.10 |
HSPA6 |
heat shock 70kDa protein 6 (HSP70B') |
1384 |
0.32 |
| chr11_66383426_66383834 | 0.10 |
RBM14 |
RNA binding motif protein 14 |
423 |
0.31 |
| chr10_71991819_71992862 | 0.10 |
PPA1 |
pyrophosphatase (inorganic) 1 |
820 |
0.66 |
| chr1_47060765_47060916 | 0.10 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
109 |
0.96 |
| chr16_70456934_70457085 | 0.10 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
15982 |
0.11 |
| chr1_212781685_212782134 | 0.10 |
ATF3 |
activating transcription factor 3 |
103 |
0.96 |
| chr6_144671797_144672109 | 0.10 |
UTRN |
utrophin |
6716 |
0.26 |
| chr17_60970209_60970370 | 0.10 |
ENSG00000207552 |
. |
51287 |
0.14 |
| chr1_202977767_202977918 | 0.10 |
TMEM183A |
transmembrane protein 183A |
1328 |
0.33 |
| chr22_22020368_22020729 | 0.10 |
PPIL2 |
peptidylprolyl isomerase (cyclophilin)-like 2 |
190 |
0.88 |
| chr16_3137327_3137594 | 0.09 |
ENSG00000200204 |
. |
1316 |
0.18 |
| chr18_29542335_29542486 | 0.09 |
TRAPPC8 |
trafficking protein particle complex 8 |
19341 |
0.14 |
| chr1_63787954_63788471 | 0.09 |
RP4-792G4.2 |
|
83 |
0.88 |
| chr3_16328270_16328443 | 0.09 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
17608 |
0.16 |
| chr9_100565675_100565842 | 0.09 |
FOXE1 |
forkhead box E1 (thyroid transcription factor 2) |
49778 |
0.12 |
| chr1_98569638_98569861 | 0.09 |
ENSG00000225206 |
. |
58022 |
0.17 |
| chr14_89771641_89771801 | 0.09 |
RP11-356K23.1 |
|
44908 |
0.14 |
| chr20_62580551_62580702 | 0.09 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
1853 |
0.14 |
| chr10_111970982_111971381 | 0.09 |
MXI1 |
MAX interactor 1, dimerization protein |
1192 |
0.49 |
| chr9_24621401_24621552 | 0.09 |
IZUMO3 |
IZUMO family member 3 |
75532 |
0.13 |
| chr7_6388445_6388661 | 0.09 |
FAM220A |
family with sequence similarity 220, member A |
37 |
0.98 |
| chr16_89307892_89308107 | 0.09 |
RP11-46C24.6 |
|
12899 |
0.11 |
| chr15_101733614_101733826 | 0.09 |
CHSY1 |
chondroitin sulfate synthase 1 |
58417 |
0.11 |
| chr1_235489879_235490582 | 0.09 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
435 |
0.56 |
| chr1_244210323_244211450 | 0.09 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
3699 |
0.27 |
| chr3_154041274_154042558 | 0.09 |
DHX36 |
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
289 |
0.93 |
| chr3_30645783_30645934 | 0.09 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
2136 |
0.46 |
| chr12_116703907_116704375 | 0.09 |
ENSG00000263768 |
. |
10348 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.4 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.1 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |