Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for BACH1_NFE2_NFE2L2
Z-value: 2.46
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.11 | BTB domain and CNC homolog 1 |
|
NFE2
|
ENSG00000123405.9 | nuclear factor, erythroid 2 |
|
NFE2L2
|
ENSG00000116044.11 | nuclear factor, erythroid 2 like 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr21_30689027_30689178 | BACH1 | 10348 | 0.182548 | 0.77 | 1.5e-02 | Click! |
| chr21_30688725_30688876 | BACH1 | 10650 | 0.182087 | 0.73 | 2.4e-02 | Click! |
| chr21_30687972_30688123 | BACH1 | 10523 | 0.182960 | 0.70 | 3.7e-02 | Click! |
| chr21_30620619_30620770 | BACH1 | 50498 | 0.100138 | 0.64 | 6.1e-02 | Click! |
| chr21_30677925_30678103 | BACH1 | 490 | 0.809035 | 0.61 | 8.1e-02 | Click! |
| chr12_54690613_54690897 | NFE2 | 913 | 0.331553 | 0.93 | 3.2e-04 | Click! |
| chr12_54688592_54688942 | NFE2 | 775 | 0.396851 | 0.92 | 4.3e-04 | Click! |
| chr12_54689177_54689328 | NFE2 | 290 | 0.775354 | 0.92 | 4.5e-04 | Click! |
| chr12_54689023_54689174 | NFE2 | 444 | 0.631982 | 0.90 | 9.1e-04 | Click! |
| chr12_54693479_54693635 | NFE2 | 1242 | 0.210655 | 0.81 | 8.6e-03 | Click! |
| chr2_178127025_178127819 | NFE2L2 | 760 | 0.459348 | -0.65 | 5.8e-02 | Click! |
| chr2_178125992_178126256 | NFE2L2 | 2058 | 0.186804 | -0.48 | 1.9e-01 | Click! |
| chr2_178104885_178105235 | NFE2L2 | 7845 | 0.111307 | 0.46 | 2.2e-01 | Click! |
| chr2_178125001_178125671 | NFE2L2 | 2846 | 0.149705 | -0.36 | 3.5e-01 | Click! |
| chr2_178140372_178140777 | NFE2L2 | 10715 | 0.107506 | 0.35 | 3.5e-01 | Click! |
Activity of the BACH1_NFE2_NFE2L2 motif across conditions
Conditions sorted by the z-value of the BACH1_NFE2_NFE2L2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
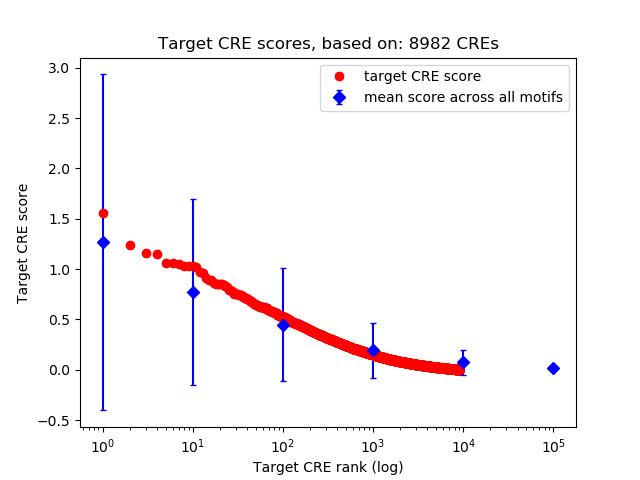
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_233249017_233249707 | 1.56 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
46691 |
0.19 |
| chr10_6443642_6443853 | 1.24 |
DKFZP667F0711 |
|
51469 |
0.16 |
| chr19_33784339_33784950 | 1.16 |
CTD-2540B15.11 |
|
6196 |
0.14 |
| chr18_47003399_47003645 | 1.15 |
RP11-110H1.4 |
|
8997 |
0.09 |
| chr22_47149775_47150260 | 1.06 |
CTA-29F11.1 |
|
8443 |
0.18 |
| chr6_147197427_147197865 | 1.06 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
25505 |
0.25 |
| chr2_111751565_111751824 | 1.05 |
ACOXL |
acyl-CoA oxidase-like |
6919 |
0.31 |
| chr19_10923780_10924057 | 1.03 |
ENSG00000207752 |
. |
4254 |
0.14 |
| chr1_10475436_10475736 | 1.03 |
APITD1 |
apoptosis-inducing, TAF9-like domain 1 |
14573 |
0.1 |
| chr7_115670709_115671033 | 1.03 |
TFEC |
transcription factor EC |
4 |
0.99 |
| chr17_74442541_74443075 | 1.02 |
UBE2O |
ubiquitin-conjugating enzyme E2O |
6480 |
0.12 |
| chr9_127520724_127520937 | 0.98 |
RP11-175D17.3 |
|
11572 |
0.14 |
| chr11_72446771_72446922 | 0.97 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
8673 |
0.12 |
| chr15_59836085_59836236 | 0.91 |
ENSG00000201704 |
. |
30261 |
0.14 |
| chr18_74189003_74189166 | 0.89 |
ZNF516 |
zinc finger protein 516 |
13651 |
0.17 |
| chr8_101231400_101231551 | 0.89 |
ENSG00000199667 |
. |
34109 |
0.13 |
| chr1_42175219_42175489 | 0.86 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
8682 |
0.27 |
| chr8_52782524_52782885 | 0.86 |
PCMTD1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
24399 |
0.15 |
| chr3_152094299_152094568 | 0.85 |
TMEM14E |
transmembrane protein 14E |
35654 |
0.17 |
| chr14_35843711_35843965 | 0.85 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
29508 |
0.2 |
| chr17_79133489_79133672 | 0.85 |
AATK-AS1 |
AATK antisense RNA 1 |
5727 |
0.1 |
| chr4_15659797_15659948 | 0.84 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
1615 |
0.4 |
| chr6_14734934_14735085 | 0.83 |
ENSG00000206960 |
. |
88243 |
0.1 |
| chr17_65427494_65427948 | 0.83 |
ENSG00000201547 |
. |
22724 |
0.12 |
| chr1_17634731_17635053 | 0.80 |
PADI4 |
peptidyl arginine deiminase, type IV |
200 |
0.93 |
| chr12_48170565_48170736 | 0.79 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
3659 |
0.16 |
| chr2_10425679_10425865 | 0.78 |
ENSG00000264030 |
. |
5015 |
0.19 |
| chr15_80452796_80453143 | 0.77 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
4676 |
0.25 |
| chr4_105982370_105982725 | 0.76 |
ENSG00000252136 |
. |
44597 |
0.16 |
| chr8_132828485_132828822 | 0.75 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
87682 |
0.1 |
| chr19_49467193_49467878 | 0.75 |
FTL |
ferritin, light polypeptide |
1023 |
0.28 |
| chr16_12910185_12910336 | 0.75 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
12386 |
0.18 |
| chr19_16567801_16568854 | 0.74 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
14427 |
0.12 |
| chr5_57555667_57556212 | 0.74 |
ENSG00000238899 |
. |
189924 |
0.03 |
| chr11_5618695_5618968 | 0.73 |
TRIM6 |
tripartite motif containing 6 |
795 |
0.34 |
| chr15_91459213_91459364 | 0.73 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
3170 |
0.11 |
| chr15_58624481_58624632 | 0.72 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
53094 |
0.15 |
| chr16_1992022_1992173 | 0.71 |
MSRB1 |
methionine sulfoxide reductase B1 |
1027 |
0.22 |
| chr16_21623104_21623255 | 0.71 |
METTL9 |
methyltransferase like 9 |
242 |
0.89 |
| chr1_43407500_43407651 | 0.71 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
10779 |
0.18 |
| chr17_441417_441568 | 0.71 |
RP5-1029F21.2 |
|
14314 |
0.17 |
| chr17_6757690_6757943 | 0.69 |
TEKT1 |
tektin 1 |
22736 |
0.14 |
| chr16_31881606_31881883 | 0.69 |
ZNF267 |
zinc finger protein 267 |
3335 |
0.29 |
| chr5_171279662_171279813 | 0.68 |
SMIM23 |
small integral membrane protein 23 |
66847 |
0.11 |
| chr2_208104248_208104487 | 0.68 |
AC007879.5 |
|
14609 |
0.21 |
| chr8_26123485_26124097 | 0.67 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
25216 |
0.23 |
| chr6_34253179_34253351 | 0.67 |
C6orf1 |
chromosome 6 open reading frame 1 |
36018 |
0.16 |
| chr17_80175660_80175811 | 0.66 |
RP13-516M14.2 |
|
3632 |
0.13 |
| chr16_18815542_18815693 | 0.65 |
RP11-1035H13.2 |
|
1212 |
0.34 |
| chr8_56814335_56814486 | 0.65 |
ENSG00000199405 |
. |
1004 |
0.46 |
| chr7_1570565_1570716 | 0.64 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
290 |
0.87 |
| chr12_56075239_56075869 | 0.64 |
METTL7B |
methyltransferase like 7B |
224 |
0.86 |
| chr21_19157038_19157189 | 0.64 |
AL109761.5 |
|
8692 |
0.24 |
| chr3_187491731_187491915 | 0.64 |
BCL6 |
B-cell CLL/lymphoma 6 |
28308 |
0.2 |
| chr1_56982304_56982688 | 0.63 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
62745 |
0.14 |
| chr20_1550154_1550305 | 0.63 |
SIRPB1 |
signal-regulatory protein beta 1 |
2329 |
0.22 |
| chr16_28607764_28607915 | 0.63 |
SULT1A2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
38 |
0.96 |
| chrX_109330942_109331348 | 0.62 |
ENSG00000265584 |
. |
5799 |
0.25 |
| chr6_13273500_13273733 | 0.62 |
PHACTR1 |
phosphatase and actin regulator 1 |
627 |
0.46 |
| chr15_94779920_94780071 | 0.62 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
5228 |
0.35 |
| chr7_44678436_44678587 | 0.62 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
14450 |
0.18 |
| chr10_115604050_115604207 | 0.62 |
DCLRE1A |
DNA cross-link repair 1A |
9731 |
0.17 |
| chr9_136710503_136710686 | 0.62 |
SARDH |
sarcosine dehydrogenase |
105517 |
0.06 |
| chr6_34632212_34632529 | 0.62 |
C6orf106 |
chromosome 6 open reading frame 106 |
7363 |
0.14 |
| chr12_124252082_124252425 | 0.61 |
DNAH10 |
dynein, axonemal, heavy chain 10 |
5211 |
0.19 |
| chr7_36636829_36636980 | 0.61 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
2642 |
0.26 |
| chr15_90420057_90420208 | 0.61 |
ENSG00000264796 |
. |
7130 |
0.11 |
| chr19_8307301_8307493 | 0.60 |
CTD-3020H12.4 |
|
2865 |
0.2 |
| chr7_1553073_1553492 | 0.59 |
AC102953.6 |
|
4219 |
0.16 |
| chr8_131012205_131012356 | 0.59 |
ENSG00000253043 |
. |
4567 |
0.2 |
| chr5_177904286_177904437 | 0.59 |
CTB-26E19.1 |
|
34266 |
0.18 |
| chr5_150439543_150439694 | 0.59 |
TNIP1 |
TNFAIP3 interacting protein 1 |
5038 |
0.22 |
| chr1_221219794_221220024 | 0.59 |
HLX |
H2.0-like homeobox |
165325 |
0.04 |
| chr2_160711950_160712101 | 0.58 |
LY75 |
lymphocyte antigen 75 |
49168 |
0.13 |
| chr16_88367140_88367361 | 0.58 |
ZNF469 |
zinc finger protein 469 |
126629 |
0.05 |
| chr6_133069776_133069927 | 0.58 |
RP1-55C23.7 |
|
3963 |
0.15 |
| chr5_39270484_39270720 | 0.58 |
FYB |
FYN binding protein |
144 |
0.98 |
| chr13_78659821_78660303 | 0.57 |
RNF219-AS1 |
RNF219 antisense RNA 1 |
30772 |
0.24 |
| chr12_51720692_51720843 | 0.57 |
BIN2 |
bridging integrator 2 |
2315 |
0.25 |
| chr16_75628443_75628594 | 0.57 |
ADAT1 |
adenosine deaminase, tRNA-specific 1 |
8514 |
0.13 |
| chr5_161696334_161696596 | 0.57 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
174925 |
0.03 |
| chr12_132345039_132345375 | 0.57 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
15367 |
0.16 |
| chr14_71165073_71165224 | 0.57 |
TTC9 |
tetratricopeptide repeat domain 9 |
56644 |
0.12 |
| chr3_149166374_149166525 | 0.56 |
TM4SF4 |
transmembrane 4 L six family member 4 |
25312 |
0.18 |
| chr10_94581825_94581976 | 0.56 |
EXOC6 |
exocyst complex component 6 |
9035 |
0.27 |
| chr19_52089706_52089857 | 0.56 |
AC018755.1 |
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
7849 |
0.12 |
| chr9_21677527_21678485 | 0.55 |
ENSG00000244230 |
. |
21307 |
0.21 |
| chr7_36789268_36789419 | 0.55 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
25189 |
0.22 |
| chr1_44693004_44693155 | 0.54 |
DMAP1 |
DNA methyltransferase 1 associated protein 1 |
13566 |
0.17 |
| chr4_83933011_83933976 | 0.54 |
LIN54 |
lin-54 homolog (C. elegans) |
547 |
0.73 |
| chr6_7697968_7698508 | 0.54 |
BMP6 |
bone morphogenetic protein 6 |
28792 |
0.23 |
| chr10_94520371_94520715 | 0.54 |
ENSG00000201412 |
. |
50178 |
0.13 |
| chr3_4793867_4794159 | 0.54 |
ENSG00000239126 |
. |
126373 |
0.05 |
| chr22_32033546_32033968 | 0.53 |
PISD |
phosphatidylserine decarboxylase |
634 |
0.74 |
| chr1_159061998_159062403 | 0.53 |
AIM2 |
absent in melanoma 2 |
15509 |
0.17 |
| chr17_41081070_41081349 | 0.53 |
ENSG00000200510 |
. |
3750 |
0.1 |
| chr5_173959609_173959903 | 0.53 |
MSX2 |
msh homeobox 2 |
191780 |
0.03 |
| chr2_39054904_39055055 | 0.53 |
DHX57 |
DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 |
8729 |
0.15 |
| chr6_140893365_140893516 | 0.53 |
ENSG00000221336 |
. |
88410 |
0.1 |
| chr16_11734508_11734659 | 0.53 |
LITAF |
lipopolysaccharide-induced TNF factor |
4346 |
0.22 |
| chr22_24906725_24906876 | 0.53 |
AP000355.2 |
|
5676 |
0.15 |
| chr16_48570933_48571121 | 0.53 |
RP11-44I10.3 |
|
22545 |
0.17 |
| chr2_161208306_161208457 | 0.52 |
ENSG00000252465 |
. |
45098 |
0.15 |
| chr5_94416178_94416653 | 0.52 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
771 |
0.78 |
| chr1_33474618_33474769 | 0.52 |
RP1-117O3.2 |
|
22017 |
0.13 |
| chr16_84766426_84766761 | 0.52 |
USP10 |
ubiquitin specific peptidase 10 |
32947 |
0.16 |
| chr20_352637_353070 | 0.52 |
TRIB3 |
tribbles pseudokinase 3 |
8408 |
0.13 |
| chr9_123446778_123446929 | 0.51 |
MEGF9 |
multiple EGF-like-domains 9 |
29759 |
0.21 |
| chr4_139162325_139164249 | 0.51 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
216 |
0.97 |
| chr19_42306064_42306215 | 0.51 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
5054 |
0.13 |
| chr8_101804090_101804262 | 0.51 |
ENSG00000202001 |
. |
36753 |
0.12 |
| chr17_25608843_25608994 | 0.51 |
ENSG00000263583 |
. |
12104 |
0.22 |
| chr20_52523665_52523816 | 0.50 |
AC005220.3 |
|
32959 |
0.21 |
| chr14_73934175_73934326 | 0.50 |
ENSG00000251393 |
. |
5121 |
0.16 |
| chr20_3993319_3993621 | 0.50 |
RNF24 |
ring finger protein 24 |
2566 |
0.31 |
| chr2_97283065_97283216 | 0.49 |
KANSL3 |
KAT8 regulatory NSL complex subunit 3 |
20906 |
0.19 |
| chr12_11898918_11899069 | 0.49 |
ETV6 |
ets variant 6 |
6442 |
0.31 |
| chr7_130527306_130527457 | 0.49 |
ENSG00000226380 |
. |
34199 |
0.2 |
| chr7_77355828_77355979 | 0.49 |
RSBN1L |
round spermatid basic protein 1-like |
29435 |
0.2 |
| chr7_65746937_65747265 | 0.49 |
TPST1 |
tyrosylprotein sulfotransferase 1 |
60510 |
0.11 |
| chr3_172035613_172035916 | 0.48 |
AC092964.1 |
Uncharacterized protein |
1546 |
0.44 |
| chr8_53372834_53372985 | 0.48 |
ST18 |
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
50544 |
0.17 |
| chr2_87868562_87868713 | 0.48 |
ENSG00000265507 |
. |
60637 |
0.14 |
| chr11_72451789_72452038 | 0.48 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
11535 |
0.12 |
| chr3_183025030_183025275 | 0.48 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
42020 |
0.14 |
| chr6_36001781_36002122 | 0.48 |
MAPK14 |
mitogen-activated protein kinase 14 |
5085 |
0.21 |
| chr3_171678813_171679061 | 0.47 |
TMEM212-IT1 |
TMEM212 intronic transcript 1 (non-protein coding) |
66711 |
0.11 |
| chr22_43288275_43288426 | 0.47 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
34238 |
0.17 |
| chrX_107683395_107684019 | 0.47 |
COL4A5 |
collagen, type IV, alpha 5 |
595 |
0.63 |
| chr15_64985334_64985485 | 0.47 |
AC100830.4 |
|
1831 |
0.21 |
| chr17_79702501_79703028 | 0.47 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
22292 |
0.07 |
| chrX_53292770_53292921 | 0.47 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
17951 |
0.16 |
| chr15_94749688_94749858 | 0.47 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
24994 |
0.27 |
| chr7_139365740_139366077 | 0.47 |
HIPK2 |
homeodomain interacting protein kinase 2 |
55933 |
0.14 |
| chr2_25979688_25979839 | 0.46 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
23415 |
0.18 |
| chr12_120669177_120669328 | 0.46 |
PXN |
paxillin |
4605 |
0.14 |
| chr1_235010269_235010484 | 0.46 |
ENSG00000239690 |
. |
29557 |
0.2 |
| chr3_10334877_10335066 | 0.46 |
GHRL |
ghrelin/obestatin prepropeptide |
340 |
0.8 |
| chr5_130976356_130976507 | 0.46 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
5502 |
0.32 |
| chr14_33883349_33883500 | 0.46 |
NPAS3 |
neuronal PAS domain protein 3 |
198906 |
0.03 |
| chr2_46689171_46689322 | 0.46 |
ENSG00000241791 |
. |
13596 |
0.16 |
| chr13_50783171_50783406 | 0.46 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
126981 |
0.05 |
| chr1_40530106_40530257 | 0.46 |
PPT1 |
palmitoyl-protein thioesterase 1 |
17812 |
0.17 |
| chr17_57807955_57808106 | 0.46 |
VMP1 |
vacuole membrane protein 1 |
954 |
0.54 |
| chr14_75165335_75165486 | 0.45 |
ENSG00000222604 |
. |
13244 |
0.13 |
| chr1_56968313_56968464 | 0.45 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
76853 |
0.11 |
| chr10_5723979_5724130 | 0.45 |
FAM208B |
family with sequence similarity 208, member B |
2747 |
0.23 |
| chr1_8682186_8682337 | 0.45 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
2178 |
0.41 |
| chr7_149318979_149319130 | 0.45 |
ZNF767 |
zinc finger family member 767 |
2602 |
0.35 |
| chr19_2169086_2169237 | 0.45 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
4977 |
0.13 |
| chr17_76871972_76872123 | 0.45 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
1815 |
0.27 |
| chr2_219020724_219020875 | 0.45 |
CXCR1 |
chemokine (C-X-C motif) receptor 1 |
10919 |
0.14 |
| chr7_23312825_23312976 | 0.44 |
MALSU1 |
mitochondrial assembly of ribosomal large subunit 1 |
25919 |
0.17 |
| chr5_39266648_39266799 | 0.44 |
FYB |
FYN binding protein |
4023 |
0.33 |
| chr18_9869922_9870073 | 0.44 |
TXNDC2 |
thioredoxin domain containing 2 (spermatozoa) |
15766 |
0.16 |
| chr19_18626356_18626507 | 0.44 |
ELL |
elongation factor RNA polymerase II |
6506 |
0.1 |
| chr4_185331100_185331251 | 0.44 |
IRF2 |
interferon regulatory factor 2 |
8569 |
0.21 |
| chr4_13959692_13959843 | 0.44 |
ENSG00000252092 |
. |
299516 |
0.01 |
| chr17_26220981_26221377 | 0.44 |
LYRM9 |
LYR motif containing 9 |
770 |
0.4 |
| chr10_125979038_125979189 | 0.44 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
125907 |
0.05 |
| chr2_71680397_71681534 | 0.44 |
DYSF |
dysferlin |
113 |
0.98 |
| chr11_78139082_78139233 | 0.44 |
GAB2 |
GRB2-associated binding protein 2 |
9763 |
0.18 |
| chr12_13145535_13145776 | 0.44 |
HEBP1 |
heme binding protein 1 |
3441 |
0.17 |
| chr17_5365675_5365826 | 0.43 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
6318 |
0.12 |
| chr5_176935395_176936034 | 0.43 |
DOK3 |
docking protein 3 |
1113 |
0.32 |
| chr2_106414710_106414861 | 0.43 |
NCK2 |
NCK adaptor protein 2 |
18230 |
0.25 |
| chr3_188301983_188302303 | 0.43 |
LPP-AS1 |
LPP antisense RNA 1 |
15689 |
0.2 |
| chr7_5481756_5481907 | 0.43 |
TNRC18 |
trinucleotide repeat containing 18 |
16786 |
0.12 |
| chr17_2060784_2060943 | 0.43 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
15609 |
0.12 |
| chr2_69809391_69809743 | 0.42 |
AAK1 |
AP2 associated kinase 1 |
61219 |
0.1 |
| chr8_48266549_48266761 | 0.42 |
SPIDR |
scaffolding protein involved in DNA repair |
86308 |
0.1 |
| chr4_40564844_40564995 | 0.42 |
RBM47 |
RNA binding motif protein 47 |
46929 |
0.14 |
| chr7_30265425_30265712 | 0.42 |
ZNRF2 |
zinc and ring finger 2 |
58355 |
0.1 |
| chr8_142274723_142274874 | 0.42 |
RP11-10J21.3 |
Uncharacterized protein |
10134 |
0.14 |
| chr2_43439997_43440275 | 0.42 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
13612 |
0.23 |
| chr20_50267277_50267428 | 0.42 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
87982 |
0.09 |
| chr15_89710840_89711060 | 0.42 |
RLBP1 |
retinaldehyde binding protein 1 |
44124 |
0.12 |
| chr6_11354405_11354556 | 0.42 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
28052 |
0.23 |
| chr8_103807477_103808006 | 0.41 |
ENSG00000266799 |
. |
61794 |
0.1 |
| chr21_30672594_30672989 | 0.41 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
332 |
0.89 |
| chr7_105316780_105317038 | 0.41 |
ATXN7L1 |
ataxin 7-like 1 |
2700 |
0.37 |
| chr2_85937073_85937224 | 0.41 |
GNLY |
granulysin |
14657 |
0.13 |
| chr1_33812781_33812932 | 0.41 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
2630 |
0.17 |
| chr2_197078455_197078606 | 0.41 |
ENSG00000239161 |
. |
1516 |
0.41 |
| chr16_87028312_87028580 | 0.41 |
RP11-899L11.3 |
|
221075 |
0.02 |
| chr11_65313728_65314148 | 0.41 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
292 |
0.8 |
| chr6_160411027_160411270 | 0.41 |
AIRN |
antisense of IGF2R non-protein coding RNA |
17548 |
0.19 |
| chrX_9320798_9321168 | 0.41 |
TBL1X |
transducin (beta)-like 1X-linked |
110352 |
0.07 |
| chr16_72085557_72085708 | 0.41 |
HP |
haptoglobin |
2859 |
0.17 |
| chr10_45870253_45870404 | 0.40 |
ALOX5 |
arachidonate 5-lipoxygenase |
653 |
0.77 |
| chr5_150863261_150863412 | 0.40 |
ENSG00000200227 |
. |
6198 |
0.17 |
| chr7_115872079_115872230 | 0.40 |
AC073130.3 |
|
7151 |
0.22 |
| chr4_99273774_99274037 | 0.40 |
RAP1GDS1 |
RAP1, GTP-GDP dissociation stimulator 1 |
26348 |
0.23 |
| chrX_13093156_13093307 | 0.40 |
FAM9C |
family with sequence similarity 9, member C |
30430 |
0.22 |
| chr10_26770074_26770395 | 0.40 |
ENSG00000199733 |
. |
28284 |
0.21 |
| chr10_89868275_89868426 | 0.40 |
ENSG00000200891 |
. |
113898 |
0.06 |
| chr2_175495703_175495854 | 0.40 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
3529 |
0.27 |
| chr10_126222901_126223052 | 0.40 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
72564 |
0.1 |
| chr7_127667480_127667631 | 0.40 |
LRRC4 |
leucine rich repeat containing 4 |
3503 |
0.31 |
| chr19_15755544_15755695 | 0.40 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
3500 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.2 | 0.9 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.2 | 0.5 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.5 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 0.2 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.1 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.1 | 0.2 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 0.1 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.1 | 0.2 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.1 | 0.2 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.1 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 0.2 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 | 0.2 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0009309 | amine biosynthetic process(GO:0009309) |
| 0.1 | 0.2 | GO:0090594 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 0.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.1 | 0.2 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.1 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.2 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.1 | 0.4 | GO:0055093 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.4 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.3 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.4 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.1 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0002902 | regulation of B cell apoptotic process(GO:0002902) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0052553 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.0 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0002830 | positive regulation of type 2 immune response(GO:0002830) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.2 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0015811 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.0 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.1 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 1.0 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.1 | GO:0047496 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.0 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0051818 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.2 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0032715 | negative regulation of interleukin-6 production(GO:0032715) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.3 | GO:0043297 | apical junction assembly(GO:0043297) |
| 0.0 | 0.0 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.0 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.4 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.2 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.1 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.3 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.3 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.6 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.5 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.2 | 0.5 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.3 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.0 | 0.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.0 | REACTOME MITOTIC M M G1 PHASES | Genes involved in Mitotic M-M/G1 phases |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |