Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for BACH2
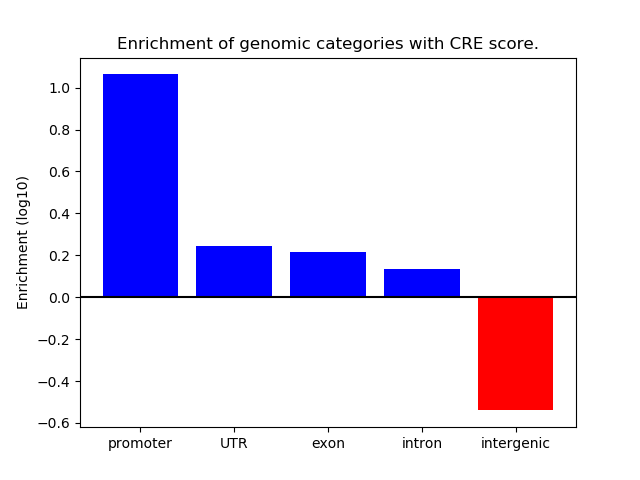
Z-value: 1.07
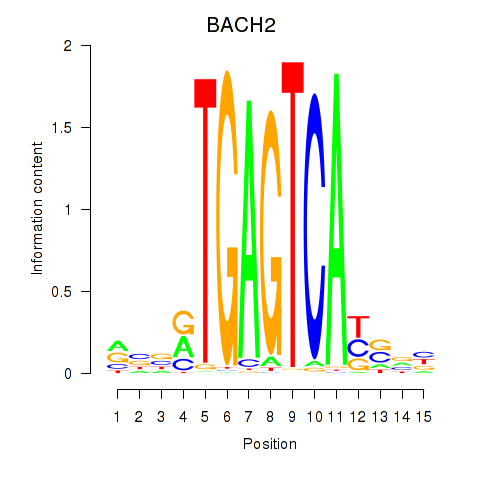
Transcription factors associated with BACH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH2
|
ENSG00000112182.10 | BTB domain and CNC homolog 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_90897115_90897402 | BACH2 | 109203 | 0.064927 | 0.76 | 1.9e-02 | Click! |
| chr6_90928649_90929126 | BACH2 | 77574 | 0.103711 | -0.74 | 2.3e-02 | Click! |
| chr6_91002405_91002556 | BACH2 | 3981 | 0.286790 | 0.73 | 2.7e-02 | Click! |
| chr6_90927892_90928130 | BACH2 | 78450 | 0.102322 | -0.64 | 6.3e-02 | Click! |
| chr6_90927639_90927889 | BACH2 | 78697 | 0.101935 | -0.62 | 7.3e-02 | Click! |
Activity of the BACH2 motif across conditions
Conditions sorted by the z-value of the BACH2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
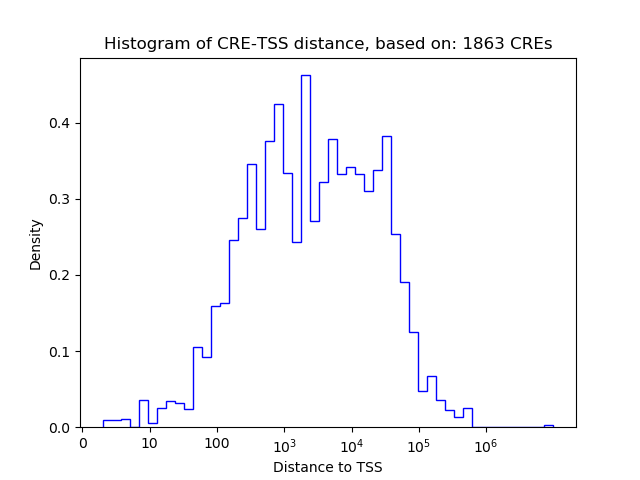
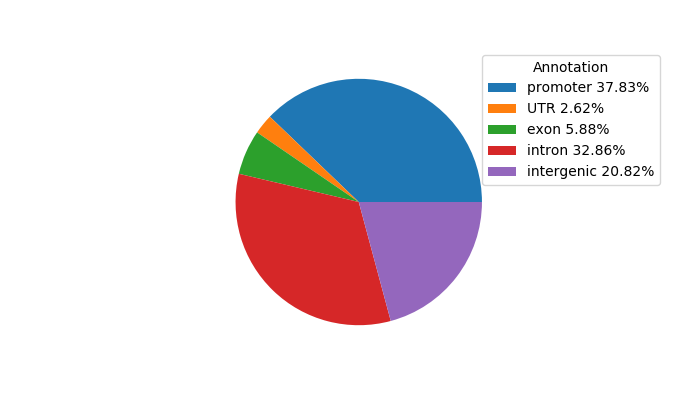
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_105531321_105531563 | 0.78 |
GPR132 |
G protein-coupled receptor 132 |
325 |
0.89 |
| chr3_196368340_196368779 | 0.66 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
1913 |
0.22 |
| chr19_2562041_2562223 | 0.64 |
CTC-265F19.2 |
|
49728 |
0.09 |
| chr5_150601304_150601455 | 0.60 |
CCDC69 |
coiled-coil domain containing 69 |
2327 |
0.29 |
| chr16_28995938_28996254 | 0.56 |
LAT |
linker for activation of T cells |
51 |
0.94 |
| chr1_45769094_45769262 | 0.52 |
HPDL |
4-hydroxyphenylpyruvate dioxygenase-like |
23367 |
0.14 |
| chr7_73508316_73508712 | 0.50 |
LIMK1 |
LIM domain kinase 1 |
1105 |
0.51 |
| chr5_130588828_130589214 | 0.50 |
CDC42SE2 |
CDC42 small effector 2 |
10681 |
0.28 |
| chr1_248069833_248069987 | 0.49 |
OR2W3 |
olfactory receptor, family 2, subfamily W, member 3 |
11021 |
0.1 |
| chr1_160983030_160983181 | 0.47 |
F11R |
F11 receptor |
7781 |
0.11 |
| chr3_47018515_47018698 | 0.45 |
CCDC12 |
coiled-coil domain containing 12 |
336 |
0.86 |
| chr1_160548517_160548751 | 0.44 |
CD84 |
CD84 molecule |
629 |
0.67 |
| chr15_77289158_77289399 | 0.44 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
1363 |
0.44 |
| chr10_6317420_6317571 | 0.44 |
ENSG00000238366 |
. |
29701 |
0.16 |
| chr7_1572516_1572667 | 0.44 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
2241 |
0.23 |
| chr6_139458021_139458384 | 0.41 |
HECA |
headcase homolog (Drosophila) |
1953 |
0.43 |
| chr2_231090336_231091061 | 0.41 |
SP140 |
SP140 nuclear body protein |
219 |
0.54 |
| chr2_28810076_28810227 | 0.41 |
PLB1 |
phospholipase B1 |
14635 |
0.23 |
| chr5_133801504_133801725 | 0.40 |
ENSG00000207222 |
. |
34361 |
0.12 |
| chr16_48642312_48642839 | 0.40 |
N4BP1 |
NEDD4 binding protein 1 |
1545 |
0.4 |
| chr12_6445436_6445689 | 0.39 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
5452 |
0.13 |
| chr18_74845425_74845585 | 0.39 |
MBP |
myelin basic protein |
705 |
0.8 |
| chr2_43448437_43448588 | 0.39 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
5236 |
0.25 |
| chr12_96592768_96592919 | 0.38 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
4451 |
0.24 |
| chr20_1470626_1470777 | 0.38 |
SIRPB2 |
signal-regulatory protein beta 2 |
1341 |
0.29 |
| chr22_46409167_46409603 | 0.37 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
36376 |
0.09 |
| chr1_31229648_31230364 | 0.37 |
LAPTM5 |
lysosomal protein transmembrane 5 |
661 |
0.67 |
| chr1_111422762_111422913 | 0.37 |
CD53 |
CD53 molecule |
7061 |
0.2 |
| chr12_57872273_57872424 | 0.36 |
ARHGAP9 |
Rho GTPase activating protein 9 |
45 |
0.94 |
| chr5_74964815_74965486 | 0.36 |
ENSG00000207333 |
. |
40274 |
0.14 |
| chr22_20830100_20830251 | 0.36 |
ENSG00000255156 |
. |
577 |
0.57 |
| chr13_50114703_50115207 | 0.35 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
26504 |
0.17 |
| chr1_26611162_26611428 | 0.35 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
4682 |
0.13 |
| chr8_126442398_126442549 | 0.34 |
TRIB1 |
tribbles pseudokinase 1 |
90 |
0.98 |
| chr6_43597230_43597731 | 0.34 |
MAD2L1BP |
MAD2L1 binding protein |
203 |
0.78 |
| chr3_47326086_47326237 | 0.34 |
KIF9 |
kinesin family member 9 |
1220 |
0.37 |
| chr17_75439465_75439616 | 0.34 |
ENSG00000200651 |
. |
1349 |
0.37 |
| chr17_75138230_75138411 | 0.33 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
852 |
0.63 |
| chr5_132389617_132389768 | 0.33 |
HSPA4 |
heat shock 70kDa protein 4 |
2038 |
0.31 |
| chr10_22919722_22919988 | 0.33 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
39213 |
0.21 |
| chr11_9385409_9385962 | 0.32 |
IPO7 |
importin 7 |
20484 |
0.16 |
| chr15_52313401_52313552 | 0.32 |
MAPK6 |
mitogen-activated protein kinase 6 |
2078 |
0.27 |
| chr20_3725549_3725774 | 0.31 |
HSPA12B |
heat shock 70kD protein 12B |
12303 |
0.12 |
| chr19_14896102_14896264 | 0.31 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
6830 |
0.15 |
| chr1_226897694_226897845 | 0.30 |
ITPKB |
inositol-trisphosphate 3-kinase B |
27390 |
0.17 |
| chr2_192053899_192054070 | 0.30 |
STAT4 |
signal transducer and activator of transcription 4 |
37662 |
0.16 |
| chr17_74495503_74495654 | 0.30 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
1875 |
0.2 |
| chr22_36781766_36782330 | 0.30 |
MYH9 |
myosin, heavy chain 9, non-muscle |
2015 |
0.32 |
| chr1_198610920_198611071 | 0.30 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
2703 |
0.34 |
| chr1_244486436_244486726 | 0.30 |
C1orf100 |
chromosome 1 open reading frame 100 |
29356 |
0.21 |
| chr1_6086993_6087271 | 0.30 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
225 |
0.93 |
| chr19_39894109_39894559 | 0.29 |
ZFP36 |
ZFP36 ring finger protein |
3119 |
0.1 |
| chr19_11449251_11449402 | 0.29 |
RAB3D |
RAB3D, member RAS oncogene family |
1018 |
0.31 |
| chr11_18405338_18405489 | 0.29 |
ENSG00000264603 |
. |
3921 |
0.14 |
| chr5_67730038_67730311 | 0.29 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
141778 |
0.05 |
| chr12_47605032_47605192 | 0.29 |
PCED1B |
PC-esterase domain containing 1B |
4940 |
0.24 |
| chrX_71321072_71321371 | 0.28 |
RGAG4 |
retrotransposon gag domain containing 4 |
30457 |
0.14 |
| chr19_909296_909447 | 0.28 |
R3HDM4 |
R3H domain containing 4 |
3860 |
0.1 |
| chr8_126308914_126309065 | 0.28 |
ENSG00000242170 |
. |
26143 |
0.21 |
| chrX_49023399_49023590 | 0.28 |
MAGIX |
MAGI family member, X-linked |
2612 |
0.12 |
| chr16_279128_279418 | 0.28 |
LUC7L |
LUC7-like (S. cerevisiae) |
148 |
0.91 |
| chr14_68717009_68717160 | 0.28 |
ENSG00000243546 |
. |
13828 |
0.27 |
| chr1_6844402_6844649 | 0.27 |
RP11-312B8.1 |
|
378 |
0.72 |
| chr2_99069202_99069353 | 0.27 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
7864 |
0.24 |
| chr7_45069535_45069723 | 0.27 |
CCM2 |
cerebral cavernous malformation 2 |
2358 |
0.23 |
| chr8_30513601_30513752 | 0.27 |
GTF2E2 |
general transcription factor IIE, polypeptide 2, beta 34kDa |
846 |
0.59 |
| chrX_41193502_41193753 | 0.27 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
212 |
0.93 |
| chr15_78374826_78374977 | 0.27 |
SH2D7 |
SH2 domain containing 7 |
4751 |
0.14 |
| chr10_73532045_73532374 | 0.27 |
C10orf54 |
chromosome 10 open reading frame 54 |
1046 |
0.53 |
| chrX_70402024_70402577 | 0.26 |
RP5-1091N2.9 |
|
15725 |
0.12 |
| chr19_3178230_3178381 | 0.26 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
431 |
0.74 |
| chr22_27054974_27055244 | 0.26 |
CRYBA4 |
crystallin, beta A4 |
37181 |
0.15 |
| chr10_45870440_45870591 | 0.26 |
ALOX5 |
arachidonate 5-lipoxygenase |
840 |
0.69 |
| chr2_99131735_99131886 | 0.26 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
4702 |
0.28 |
| chr1_154378708_154379562 | 0.26 |
RP11-350G8.5 |
|
95 |
0.93 |
| chr12_14924776_14925101 | 0.26 |
HIST4H4 |
histone cluster 4, H4 |
873 |
0.43 |
| chrY_15016828_15017148 | 0.25 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
246 |
0.96 |
| chr9_36141464_36141615 | 0.25 |
GLIPR2 |
GLI pathogenesis-related 2 |
4797 |
0.21 |
| chr14_75085047_75085411 | 0.25 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
5923 |
0.21 |
| chr13_41884953_41885353 | 0.25 |
NAA16 |
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
188 |
0.95 |
| chr3_5055062_5055354 | 0.25 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
33562 |
0.15 |
| chr8_67592155_67592325 | 0.25 |
C8orf44 |
chromosome 8 open reading frame 44 |
3786 |
0.17 |
| chr17_56410108_56410593 | 0.25 |
MIR142 |
microRNA 142 |
481 |
0.66 |
| chr19_48246517_48246668 | 0.24 |
GLTSCR2 |
glioma tumor suppressor candidate region gene 2 |
2187 |
0.21 |
| chr17_20601796_20602095 | 0.24 |
AC126365.1 |
|
17851 |
0.23 |
| chr19_2089959_2090185 | 0.24 |
MOB3A |
MOB kinase activator 3A |
103 |
0.94 |
| chr1_95261518_95261669 | 0.24 |
SLC44A3 |
solute carrier family 44, member 3 |
24305 |
0.19 |
| chr18_3595540_3596159 | 0.24 |
DLGAP1-AS1 |
DLGAP1 antisense RNA 1 |
1396 |
0.36 |
| chr1_40157482_40157633 | 0.24 |
HPCAL4 |
hippocalcin like 4 |
196 |
0.61 |
| chr1_36852243_36852394 | 0.24 |
STK40 |
serine/threonine kinase 40 |
821 |
0.5 |
| chr12_1434091_1434389 | 0.24 |
RP5-951N9.2 |
|
60759 |
0.12 |
| chr7_5595689_5595960 | 0.24 |
CTB-161C1.1 |
|
538 |
0.69 |
| chr2_122288534_122288940 | 0.24 |
ENSG00000264229 |
. |
280 |
0.79 |
| chr10_45870253_45870404 | 0.23 |
ALOX5 |
arachidonate 5-lipoxygenase |
653 |
0.77 |
| chr11_47787507_47787658 | 0.23 |
FNBP4 |
formin binding protein 4 |
1271 |
0.36 |
| chr13_75900551_75900966 | 0.23 |
TBC1D4 |
TBC1 domain family, member 4 |
14909 |
0.24 |
| chrX_153941197_153941348 | 0.23 |
GAB3 |
GRB2-associated binding protein 3 |
32776 |
0.1 |
| chr22_32871360_32872181 | 0.23 |
FBXO7 |
F-box protein 7 |
190 |
0.94 |
| chr11_118754601_118754752 | 0.23 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
201 |
0.88 |
| chr14_103801113_103802028 | 0.23 |
EIF5 |
eukaryotic translation initiation factor 5 |
284 |
0.87 |
| chr2_47649361_47649512 | 0.23 |
MSH2 |
mutS homolog 2 |
19167 |
0.17 |
| chr3_50354430_50354581 | 0.23 |
HYAL2 |
hyaluronoglucosaminidase 2 |
3874 |
0.08 |
| chr2_114008714_114008865 | 0.23 |
ENSG00000189223 |
. |
446 |
0.78 |
| chr19_47853304_47853535 | 0.23 |
DHX34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
881 |
0.52 |
| chr19_14494809_14495344 | 0.23 |
CD97 |
CD97 molecule |
2820 |
0.2 |
| chr1_223738388_223738539 | 0.23 |
CAPN8 |
calpain 8 |
14083 |
0.27 |
| chr13_34116508_34116659 | 0.23 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
191816 |
0.03 |
| chr8_145022714_145022952 | 0.23 |
PLEC |
plectin |
2118 |
0.18 |
| chr20_57721681_57721954 | 0.23 |
ZNF831 |
zinc finger protein 831 |
44258 |
0.15 |
| chr12_53616534_53616822 | 0.23 |
RARG |
retinoic acid receptor, gamma |
2481 |
0.17 |
| chr15_89952076_89952330 | 0.23 |
ENSG00000207819 |
. |
40955 |
0.13 |
| chr12_15104110_15104261 | 0.22 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
100 |
0.96 |
| chr17_44257050_44257201 | 0.22 |
KANSL1 |
KAT8 regulatory NSL complex subunit 1 |
7531 |
0.17 |
| chr3_32995983_32996275 | 0.22 |
CCR4 |
chemokine (C-C motif) receptor 4 |
3063 |
0.34 |
| chr12_109022383_109022629 | 0.22 |
RP11-689B22.2 |
|
43 |
0.96 |
| chr18_43754040_43754305 | 0.22 |
C18orf25 |
chromosome 18 open reading frame 25 |
172 |
0.96 |
| chr14_104094277_104094428 | 0.22 |
KLC1 |
kinesin light chain 1 |
1162 |
0.33 |
| chr4_38762729_38763007 | 0.22 |
ENSG00000222230 |
. |
2355 |
0.26 |
| chr9_130717791_130717942 | 0.22 |
FAM102A |
family with sequence similarity 102, member A |
4865 |
0.11 |
| chr17_78865183_78865610 | 0.22 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
31128 |
0.13 |
| chr1_158789498_158789649 | 0.22 |
MNDA |
myeloid cell nuclear differentiation antigen |
11534 |
0.17 |
| chr1_45196821_45197082 | 0.21 |
ENSG00000200169 |
. |
109 |
0.92 |
| chr1_28640614_28640765 | 0.21 |
MED18 |
mediator complex subunit 18 |
14860 |
0.15 |
| chr3_190304394_190304671 | 0.21 |
IL1RAP |
interleukin 1 receptor accessory protein |
23297 |
0.23 |
| chr16_67571099_67571332 | 0.21 |
FAM65A |
family with sequence similarity 65, member A |
142 |
0.9 |
| chr9_132595482_132595633 | 0.21 |
C9orf78 |
chromosome 9 open reading frame 78 |
1997 |
0.23 |
| chr15_89630929_89631312 | 0.21 |
RP11-326A19.3 |
|
52 |
0.77 |
| chr5_177540474_177540824 | 0.21 |
N4BP3 |
NEDD4 binding protein 3 |
205 |
0.93 |
| chr20_33680803_33681037 | 0.21 |
TRPC4AP |
transient receptor potential cation channel, subfamily C, member 4 associated protein |
246 |
0.9 |
| chr5_169705883_169706422 | 0.21 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
11821 |
0.22 |
| chr9_117134390_117135168 | 0.20 |
AKNA |
AT-hook transcription factor |
4465 |
0.23 |
| chr6_35568962_35570321 | 0.20 |
ENSG00000212579 |
. |
49954 |
0.1 |
| chr5_10632325_10632658 | 0.20 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
4154 |
0.28 |
| chr1_8000859_8001010 | 0.20 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
8 |
0.98 |
| chr5_167229311_167229462 | 0.20 |
TENM2 |
teneurin transmembrane protein 2 |
47378 |
0.15 |
| chr12_120669177_120669328 | 0.20 |
PXN |
paxillin |
4605 |
0.14 |
| chr13_108919519_108919670 | 0.20 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
2383 |
0.32 |
| chr1_172628190_172628341 | 0.20 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
107 |
0.98 |
| chr2_64870286_64870742 | 0.20 |
SERTAD2 |
SERTA domain containing 2 |
10533 |
0.23 |
| chr19_51920344_51920879 | 0.20 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
224 |
0.81 |
| chr14_94425760_94425911 | 0.20 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
2068 |
0.26 |
| chr3_101655473_101655624 | 0.20 |
ENSG00000202177 |
. |
18373 |
0.23 |
| chr22_43288275_43288426 | 0.20 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
34238 |
0.17 |
| chr18_9404902_9405053 | 0.20 |
RALBP1 |
ralA binding protein 1 |
70030 |
0.08 |
| chr17_45814129_45814344 | 0.20 |
TBX21 |
T-box 21 |
3626 |
0.18 |
| chr17_34409877_34410116 | 0.20 |
AC069363.1 |
|
1004 |
0.35 |
| chr19_13957589_13958095 | 0.19 |
ENSG00000207980 |
. |
10369 |
0.08 |
| chr2_99082787_99082938 | 0.19 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
21449 |
0.22 |
| chr20_45983820_45984266 | 0.19 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
358 |
0.81 |
| chr1_954607_954781 | 0.19 |
AGRN |
agrin |
809 |
0.4 |
| chr16_31276706_31276857 | 0.19 |
ENSG00000252876 |
. |
2031 |
0.19 |
| chr2_118943251_118943914 | 0.19 |
INSIG2 |
insulin induced gene 2 |
97532 |
0.08 |
| chr1_109791733_109792083 | 0.19 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
733 |
0.6 |
| chr10_11726764_11728101 | 0.19 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
56933 |
0.13 |
| chr16_25060015_25060182 | 0.19 |
ARHGAP17 |
Rho GTPase activating protein 17 |
33111 |
0.18 |
| chr16_6522320_6522471 | 0.19 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
10785 |
0.3 |
| chr3_72226253_72226548 | 0.19 |
ENSG00000212070 |
. |
85179 |
0.1 |
| chr3_186993064_186993215 | 0.19 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
16346 |
0.21 |
| chr6_34283894_34284092 | 0.19 |
C6orf1 |
chromosome 6 open reading frame 1 |
66746 |
0.09 |
| chr1_10570811_10571317 | 0.19 |
PEX14 |
peroxisomal biogenesis factor 14 |
36025 |
0.11 |
| chr19_39321682_39322469 | 0.19 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
224 |
0.82 |
| chr4_8230456_8230607 | 0.19 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
12040 |
0.22 |
| chr10_102758058_102758214 | 0.19 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
31 |
0.95 |
| chr4_14198140_14198291 | 0.18 |
ENSG00000202449 |
. |
494119 |
0.0 |
| chr20_62368732_62368892 | 0.18 |
LIME1 |
Lck interacting transmembrane adaptor 1 |
175 |
0.82 |
| chr8_54571850_54572001 | 0.18 |
ATP6V1H |
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
180596 |
0.03 |
| chr15_61013207_61013362 | 0.18 |
ENSG00000212625 |
. |
15684 |
0.2 |
| chr8_59466247_59466777 | 0.18 |
SDCBP |
syndecan binding protein (syntenin) |
698 |
0.76 |
| chr1_2266428_2266579 | 0.18 |
AL589739.1 |
Uncharacterized protein |
7922 |
0.11 |
| chr6_135408926_135409077 | 0.18 |
HBS1L |
HBS1-like (S. cerevisiae) |
15193 |
0.21 |
| chr12_120690170_120690321 | 0.18 |
PXN |
paxillin |
2281 |
0.2 |
| chr20_30160458_30160983 | 0.18 |
HM13-AS1 |
HM13 antisense RNA 1 |
346 |
0.8 |
| chr11_123985768_123986166 | 0.18 |
VWA5A |
von Willebrand factor A domain containing 5A |
102 |
0.97 |
| chr18_9825591_9825815 | 0.18 |
ENSG00000242651 |
. |
4921 |
0.21 |
| chr3_30653355_30653657 | 0.18 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
5413 |
0.33 |
| chr17_79651117_79651428 | 0.18 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
134 |
0.69 |
| chr17_61904834_61905197 | 0.18 |
FTSJ3 |
FtsJ homolog 3 (E. coli) |
8 |
0.53 |
| chr20_61455814_61456027 | 0.18 |
COL9A3 |
collagen, type IX, alpha 3 |
6757 |
0.12 |
| chrX_13665897_13666048 | 0.18 |
TCEANC |
transcription elongation factor A (SII) N-terminal and central domain containing |
5253 |
0.22 |
| chr17_1552617_1553136 | 0.18 |
RILP |
Rab interacting lysosomal protein |
495 |
0.61 |
| chr11_33912969_33913137 | 0.18 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
783 |
0.67 |
| chr1_235668084_235668715 | 0.18 |
B3GALNT2 |
beta-1,3-N-acetylgalactosaminyltransferase 2 |
618 |
0.8 |
| chrX_56820275_56820426 | 0.18 |
ENSG00000204272 |
. |
64658 |
0.14 |
| chr16_30077190_30078018 | 0.18 |
ALDOA |
aldolase A, fructose-bisphosphate |
96 |
0.92 |
| chr18_2658233_2658442 | 0.18 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
2451 |
0.24 |
| chr9_130681751_130681902 | 0.18 |
ST6GALNAC4 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
2521 |
0.13 |
| chr4_40201423_40201705 | 0.18 |
RHOH |
ras homolog family member H |
400 |
0.87 |
| chr22_37625645_37625816 | 0.18 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
14558 |
0.13 |
| chr5_140090759_140091080 | 0.18 |
ENSG00000199990 |
. |
59 |
0.93 |
| chr10_6019287_6019858 | 0.17 |
IL15RA |
interleukin 15 receptor, alpha |
2 |
0.97 |
| chr10_8095504_8096368 | 0.17 |
GATA3 |
GATA binding protein 3 |
720 |
0.81 |
| chr11_104914891_104915295 | 0.17 |
CARD16 |
caspase recruitment domain family, member 16 |
941 |
0.54 |
| chr5_42950238_42951303 | 0.17 |
SEPP1 |
selenoprotein P, plasma, 1 |
63276 |
0.1 |
| chr20_44439937_44440088 | 0.17 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
1203 |
0.27 |
| chr3_37033296_37034262 | 0.17 |
EPM2AIP1 |
EPM2A (laforin) interacting protein 1 |
1016 |
0.36 |
| chr1_202777207_202777594 | 0.17 |
KDM5B |
lysine (K)-specific demethylase 5B |
146 |
0.92 |
| chr8_125384746_125384961 | 0.17 |
TMEM65 |
transmembrane protein 65 |
80 |
0.98 |
| chrX_19816618_19816769 | 0.17 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
1176 |
0.63 |
| chr13_28000222_28001106 | 0.17 |
GTF3A |
general transcription factor IIIA |
1983 |
0.29 |
| chr17_80817564_80818630 | 0.17 |
TBCD |
tubulin folding cofactor D |
173 |
0.94 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.6 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.3 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0034243 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0002866 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) positive regulation of acute inflammatory response to antigenic stimulus(GO:0002866) positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.0 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0060896 | neural plate pattern specification(GO:0060896) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.2 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |