Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
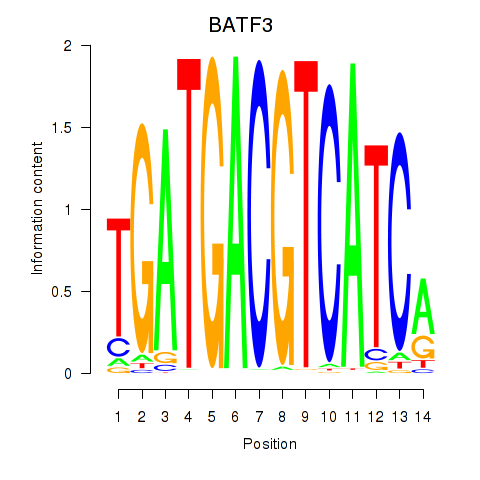
Results for BATF3
Z-value: 0.19
Transcription factors associated with BATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BATF3
|
ENSG00000123685.4 | basic leucine zipper ATF-like transcription factor 3 |
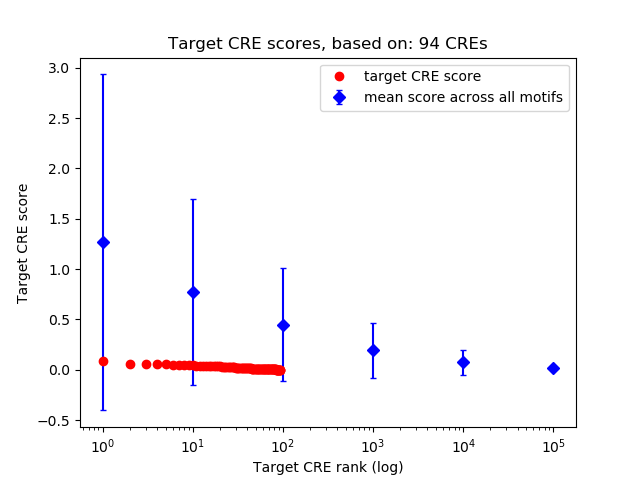
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_212873408_212874176 | BATF3 | 465 | 0.794313 | 0.54 | 1.3e-01 | Click! |
| chr1_212872713_212873036 | BATF3 | 453 | 0.800510 | 0.44 | 2.4e-01 | Click! |
| chr1_212872305_212872712 | BATF3 | 819 | 0.590846 | 0.34 | 3.7e-01 | Click! |
| chr1_212894826_212895102 | BATF3 | 21637 | 0.152140 | 0.32 | 4.0e-01 | Click! |
| chr1_212874293_212874444 | BATF3 | 1041 | 0.497527 | 0.25 | 5.1e-01 | Click! |
Activity of the BATF3 motif across conditions
Conditions sorted by the z-value of the BATF3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
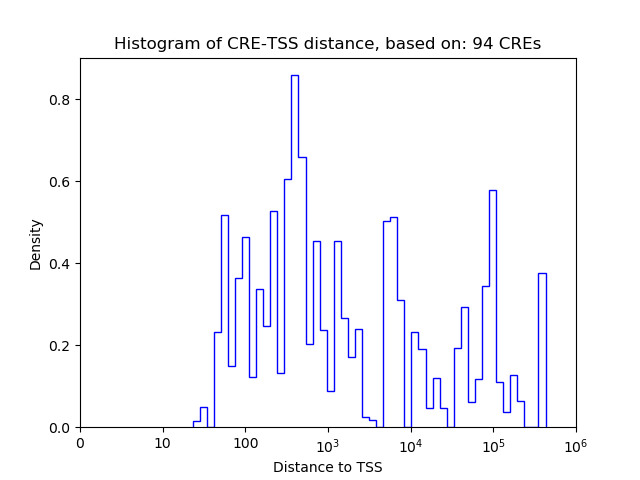
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_228353838_228353989 | 0.09 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
397 |
0.6 |
| chr6_82569789_82569940 | 0.06 |
ENSG00000206886 |
. |
96123 |
0.08 |
| chr19_8454704_8454922 | 0.06 |
RAB11B |
RAB11B, member RAS oncogene family |
52 |
0.65 |
| chr2_8144573_8144775 | 0.06 |
ENSG00000221255 |
. |
427702 |
0.01 |
| chr14_102785719_102785950 | 0.05 |
ZNF839 |
zinc finger protein 839 |
199 |
0.92 |
| chr22_39745318_39745552 | 0.05 |
SYNGR1 |
synaptogyrin 1 |
495 |
0.71 |
| chr1_198904042_198904315 | 0.05 |
ENSG00000207759 |
. |
75896 |
0.11 |
| chr12_7282858_7283009 | 0.05 |
RP11-273B20.1 |
|
85 |
0.56 |
| chr4_170580994_170582031 | 0.05 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
299 |
0.93 |
| chr5_176881613_176881764 | 0.04 |
PRR7 |
proline rich 7 (synaptic) |
6353 |
0.1 |
| chr6_27832819_27832970 | 0.04 |
HIST1H2AL |
histone cluster 1, H2al |
140 |
0.87 |
| chr13_50698385_50698672 | 0.04 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
42221 |
0.15 |
| chr4_88930112_88930263 | 0.04 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
1367 |
0.43 |
| chr5_112849172_112849403 | 0.04 |
YTHDC2 |
YTH domain containing 2 |
93 |
0.97 |
| chr12_10771207_10771388 | 0.04 |
MAGOHB |
mago-nashi homolog B (Drosophila) |
5075 |
0.18 |
| chr22_51059513_51059819 | 0.04 |
ARSA |
arylsulfatase A |
6917 |
0.1 |
| chr22_21333840_21334133 | 0.04 |
LZTR1 |
leucine-zipper-like transcription regulator 1 |
2316 |
0.14 |
| chr19_2775817_2776209 | 0.03 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
5234 |
0.12 |
| chr5_33441537_33442301 | 0.03 |
TARS |
threonyl-tRNA synthetase |
432 |
0.78 |
| chr3_159571158_159571312 | 0.03 |
SCHIP1 |
schwannomin interacting protein 1 |
507 |
0.8 |
| chrX_40943933_40944084 | 0.03 |
USP9X |
ubiquitin specific peptidase 9, X-linked |
880 |
0.73 |
| chr3_101405604_101405866 | 0.03 |
RPL24 |
ribosomal protein L24 |
109 |
0.94 |
| chr16_67564286_67564635 | 0.03 |
FAM65A |
family with sequence similarity 65, member A |
766 |
0.44 |
| chr22_22020368_22020729 | 0.03 |
PPIL2 |
peptidylprolyl isomerase (cyclophilin)-like 2 |
190 |
0.88 |
| chr14_100112036_100112187 | 0.03 |
HHIPL1 |
HHIP-like 1 |
631 |
0.72 |
| chr17_34115736_34115943 | 0.03 |
MMP28 |
matrix metallopeptidase 28 |
6872 |
0.1 |
| chr12_82794791_82794942 | 0.02 |
METTL25 |
methyltransferase like 25 |
1728 |
0.44 |
| chr1_234176768_234176919 | 0.02 |
RP11-488L4.1 |
|
89990 |
0.09 |
| chr7_151575210_151575378 | 0.02 |
PRKAG2-AS1 |
PRKAG2 antisense RNA 1 |
744 |
0.48 |
| chr13_36871577_36872517 | 0.02 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
68 |
0.34 |
| chr8_21967077_21967500 | 0.02 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
356 |
0.79 |
| chr17_38560668_38560819 | 0.02 |
ENSG00000222881 |
. |
13153 |
0.11 |
| chr1_168147799_168148070 | 0.02 |
TIPRL |
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
237 |
0.94 |
| chr21_46898660_46898949 | 0.02 |
COL18A1 |
collagen, type XVIII, alpha 1 |
11385 |
0.19 |
| chr13_32525396_32525547 | 0.02 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
1246 |
0.59 |
| chr6_3157575_3157860 | 0.02 |
TUBB2A |
tubulin, beta 2A class IIa |
43 |
0.97 |
| chr18_29542335_29542486 | 0.02 |
TRAPPC8 |
trafficking protein particle complex 8 |
19341 |
0.14 |
| chr10_99496753_99496904 | 0.02 |
ZFYVE27 |
zinc finger, FYVE domain containing 27 |
50 |
0.97 |
| chr12_25486836_25487071 | 0.02 |
ENSG00000201439 |
. |
70409 |
0.1 |
| chr3_28877353_28877504 | 0.02 |
ENSG00000238470 |
. |
175305 |
0.03 |
| chr11_108535331_108535645 | 0.02 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
264 |
0.95 |
| chr22_36519910_36520146 | 0.02 |
APOL3 |
apolipoprotein L, 3 |
36803 |
0.17 |
| chr20_62580551_62580702 | 0.01 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
1853 |
0.14 |
| chr14_100028328_100028479 | 0.01 |
CCDC85C |
coiled-coil domain containing 85C |
10909 |
0.17 |
| chr2_202645711_202646712 | 0.01 |
ALS2 |
amyotrophic lateral sclerosis 2 (juvenile) |
299 |
0.88 |
| chr6_91006489_91006667 | 0.01 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
49 |
0.98 |
| chr1_22379888_22380679 | 0.01 |
CDC42 |
cell division cycle 42 |
493 |
0.71 |
| chr13_97876069_97876689 | 0.01 |
MBNL2 |
muscleblind-like splicing regulator 2 |
1770 |
0.5 |
| chr3_115231729_115232383 | 0.01 |
GAP43 |
growth associated protein 43 |
110115 |
0.07 |
| chr1_26496320_26496791 | 0.01 |
ZNF593 |
zinc finger protein 593 |
122 |
0.88 |
| chr1_40860813_40861098 | 0.01 |
SMAP2 |
small ArfGAP2 |
1552 |
0.37 |
| chr2_19774725_19774876 | 0.01 |
OSR1 |
odd-skipped related transciption factor 1 |
216386 |
0.02 |
| chr5_176569173_176569324 | 0.01 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
7143 |
0.21 |
| chr15_101733614_101733826 | 0.01 |
CHSY1 |
chondroitin sulfate synthase 1 |
58417 |
0.11 |
| chr1_16677480_16677631 | 0.01 |
FBXO42 |
F-box protein 42 |
1394 |
0.38 |
| chr12_117256876_117257597 | 0.01 |
ENSG00000201382 |
. |
37326 |
0.16 |
| chr2_220109752_220110249 | 0.01 |
GLB1L |
galactosidase, beta 1-like |
128 |
0.54 |
| chr15_72766100_72766509 | 0.01 |
ARIH1 |
ariadne RBR E3 ubiquitin protein ligase 1 |
363 |
0.89 |
| chr20_42219265_42219562 | 0.01 |
IFT52 |
intraflagellar transport 52 homolog (Chlamydomonas) |
158 |
0.95 |
| chr8_10696975_10697533 | 0.01 |
PINX1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
27 |
0.57 |
| chr9_139117671_139117822 | 0.01 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
1023 |
0.52 |
| chr1_101334241_101334590 | 0.01 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
25805 |
0.15 |
| chr2_96873223_96873536 | 0.01 |
ENSG00000204685 |
. |
775 |
0.47 |
| chr14_69328469_69328679 | 0.01 |
ENSG00000263694 |
. |
16055 |
0.2 |
| chr6_144671797_144672109 | 0.01 |
UTRN |
utrophin |
6716 |
0.26 |
| chr6_27101806_27101969 | 0.01 |
HIST1H2AG |
histone cluster 1, H2ag |
1055 |
0.31 |
| chr2_28582064_28582304 | 0.01 |
FOSL2 |
FOS-like antigen 2 |
33485 |
0.14 |
| chr8_74792637_74792788 | 0.01 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
1567 |
0.43 |
| chr2_65659623_65660251 | 0.01 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
166 |
0.97 |
| chr4_136436588_136436739 | 0.01 |
ENSG00000207849 |
. |
143827 |
0.05 |
| chr3_107646209_107646537 | 0.01 |
BBX |
bobby sox homolog (Drosophila) |
128935 |
0.06 |
| chr11_66059397_66059767 | 0.01 |
TMEM151A |
transmembrane protein 151A |
241 |
0.77 |
| chr3_53380473_53381043 | 0.00 |
DCP1A |
decapping mRNA 1A |
794 |
0.58 |
| chr1_181056884_181057035 | 0.00 |
IER5 |
immediate early response 5 |
679 |
0.74 |
| chr1_9790575_9790726 | 0.00 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
12523 |
0.17 |
| chr1_23883319_23883470 | 0.00 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
2891 |
0.25 |
| chr6_28864112_28865009 | 0.00 |
HCG14 |
HLA complex group 14 (non-protein coding) |
253 |
0.89 |
| chr19_57874813_57875444 | 0.00 |
TRAPPC2P1 |
trafficking protein particle complex 2 pseudogene 1 |
193 |
0.32 |
| chr22_50912392_50912937 | 0.00 |
SBF1 |
SET binding factor 1 |
707 |
0.46 |
| chr22_44207671_44207833 | 0.00 |
EFCAB6 |
EF-hand calcium binding domain 6 |
318 |
0.69 |
| chr10_123922760_123923208 | 0.00 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
43 |
0.99 |
| chr3_146905614_146905765 | 0.00 |
RP11-649A16.1 |
|
182754 |
0.03 |
| chr12_110010896_110011058 | 0.00 |
MVK |
mevalonate kinase |
83 |
0.79 |
| chr16_2178420_2178600 | 0.00 |
ENSG00000200059 |
. |
3342 |
0.07 |
| chr1_111746296_111746584 | 0.00 |
DENND2D |
DENN/MADD domain containing 2D |
591 |
0.63 |
| chr6_31939259_31939914 | 0.00 |
STK19 |
serine/threonine kinase 19 |
22 |
0.66 |
| chr1_151445466_151445952 | 0.00 |
POGZ |
pogo transposable element with ZNF domain |
13768 |
0.11 |
| chrX_155111070_155111694 | 0.00 |
VAMP7 |
vesicle-associated membrane protein 7 |
341 |
0.92 |
| chr17_19367994_19368145 | 0.00 |
ENSG00000265335 |
. |
44477 |
0.09 |
| chr22_25815184_25815434 | 0.00 |
LRP5L |
low density lipoprotein receptor-related protein 5-like |
13965 |
0.17 |
| chr7_27232397_27232921 | 0.00 |
HOXA11-AS |
HOXA11 antisense RNA |
5221 |
0.07 |
| chr5_138739039_138739512 | 0.00 |
SPATA24 |
spermatogenesis associated 24 |
464 |
0.67 |
| chr3_151090226_151090493 | 0.00 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
12241 |
0.19 |
| chr19_18723621_18724041 | 0.00 |
TMEM59L |
transmembrane protein 59-like |
149 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |