Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
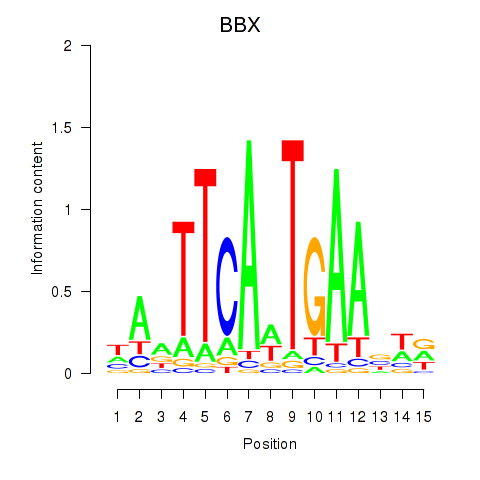
Results for BBX
Z-value: 1.79
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.14 | BBX high mobility group box domain containing |
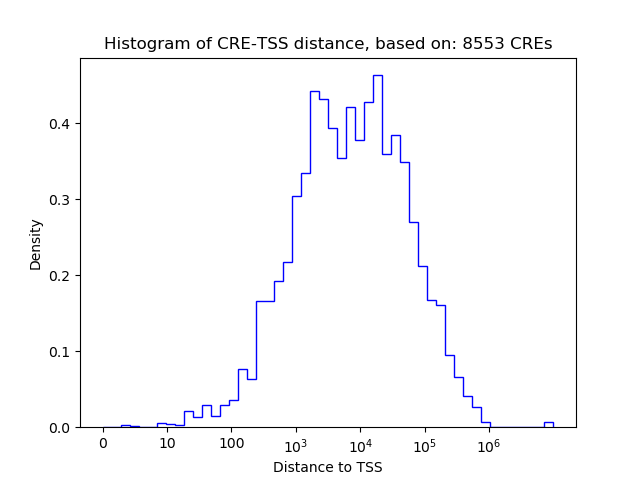
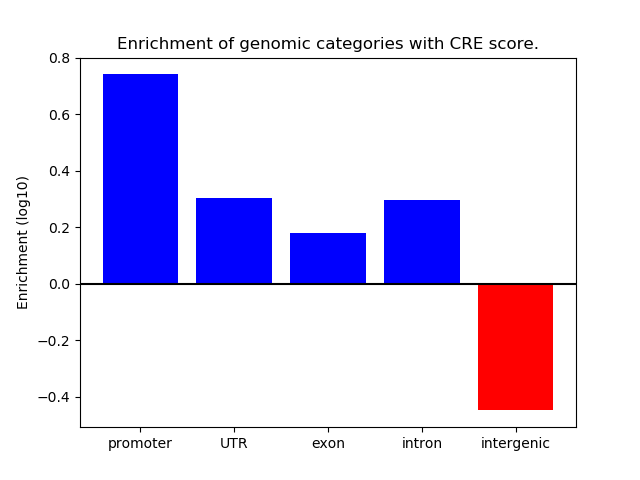
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_107381179_107381566 | BBX | 64 | 0.987673 | -0.66 | 5.1e-02 | Click! |
| chr3_107244861_107245089 | BBX | 726 | 0.804364 | -0.51 | 1.6e-01 | Click! |
| chr3_107243761_107243927 | BBX | 394 | 0.921629 | -0.50 | 1.7e-01 | Click! |
| chr3_107564740_107564985 | BBX | 47424 | 0.197769 | -0.50 | 1.7e-01 | Click! |
| chr3_107380658_107381123 | BBX | 546 | 0.876237 | -0.49 | 1.8e-01 | Click! |
Activity of the BBX motif across conditions
Conditions sorted by the z-value of the BBX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
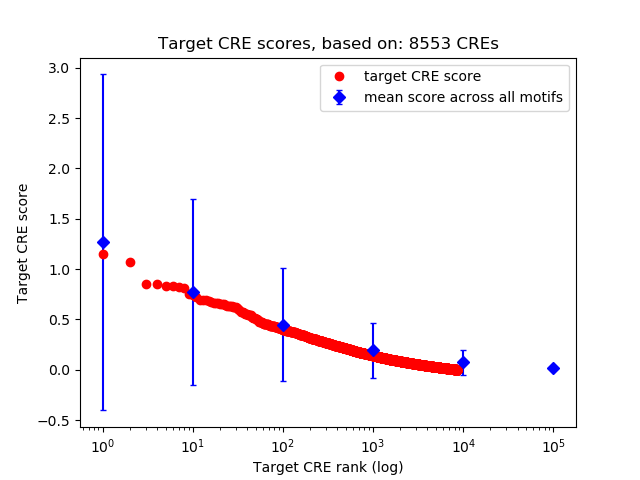
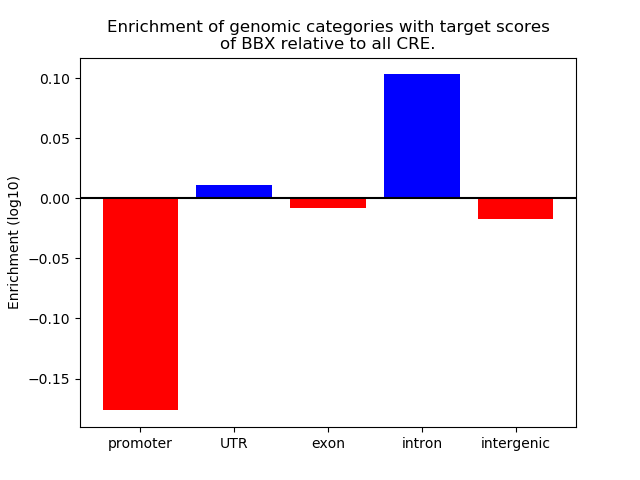
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_148189087_148189390 | 1.15 |
ADRB2 |
adrenoceptor beta 2, surface |
16918 |
0.25 |
| chr2_143887583_143887860 | 1.07 |
ARHGAP15 |
Rho GTPase activating protein 15 |
838 |
0.72 |
| chr3_16329316_16329467 | 0.85 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
18643 |
0.15 |
| chr8_133771865_133772108 | 0.85 |
TMEM71 |
transmembrane protein 71 |
808 |
0.63 |
| chr12_15109499_15109902 | 0.83 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
4500 |
0.19 |
| chr13_99957828_99957979 | 0.83 |
GPR183 |
G protein-coupled receptor 183 |
1756 |
0.38 |
| chr10_6103670_6104125 | 0.82 |
IL2RA |
interleukin 2 receptor, alpha |
356 |
0.84 |
| chr2_235362717_235362976 | 0.81 |
ARL4C |
ADP-ribosylation factor-like 4C |
42398 |
0.22 |
| chr7_150148744_150149359 | 0.75 |
GIMAP8 |
GTPase, IMAP family member 8 |
1333 |
0.41 |
| chr14_22695818_22695969 | 0.74 |
ENSG00000238634 |
. |
85006 |
0.09 |
| chr16_81031171_81031400 | 0.73 |
CMC2 |
C-x(9)-C motif containing 2 |
979 |
0.45 |
| chr7_156761668_156761819 | 0.69 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
19326 |
0.16 |
| chr7_150217655_150217806 | 0.69 |
GIMAP7 |
GTPase, IMAP family member 7 |
5812 |
0.21 |
| chr13_74807691_74807851 | 0.69 |
ENSG00000206617 |
. |
55580 |
0.16 |
| chr19_2892956_2893107 | 0.69 |
ZNF57 |
zinc finger protein 57 |
7865 |
0.11 |
| chr2_196122609_196122760 | 0.67 |
ENSG00000202206 |
. |
256076 |
0.02 |
| chr21_40139305_40139870 | 0.67 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
37644 |
0.21 |
| chr7_50349443_50349694 | 0.66 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1250 |
0.61 |
| chr6_139459897_139460408 | 0.66 |
HECA |
headcase homolog (Drosophila) |
3903 |
0.31 |
| chr1_198609761_198609912 | 0.66 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
1544 |
0.48 |
| chr12_67107724_67108271 | 0.65 |
GRIP1 |
glutamate receptor interacting protein 1 |
35072 |
0.24 |
| chr17_37933203_37933446 | 0.65 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1154 |
0.4 |
| chr7_50345819_50346403 | 0.64 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1733 |
0.5 |
| chr2_68963544_68963853 | 0.64 |
ARHGAP25 |
Rho GTPase activating protein 25 |
1684 |
0.46 |
| chr20_62270981_62271132 | 0.63 |
CTD-3184A7.4 |
|
12453 |
0.09 |
| chr10_127687165_127687316 | 0.63 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
3254 |
0.27 |
| chr18_21573835_21574026 | 0.63 |
TTC39C |
tetratricopeptide repeat domain 39C |
1193 |
0.47 |
| chr1_186292481_186292673 | 0.63 |
ENSG00000202025 |
. |
11517 |
0.17 |
| chr12_4385782_4386106 | 0.62 |
CCND2-AS1 |
CCND2 antisense RNA 1 |
594 |
0.46 |
| chr10_111836950_111837776 | 0.62 |
ADD3 |
adducin 3 (gamma) |
69641 |
0.1 |
| chr7_150146102_150146542 | 0.61 |
GIMAP8 |
GTPase, IMAP family member 8 |
1396 |
0.39 |
| chr21_35305474_35305968 | 0.60 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
2203 |
0.25 |
| chr5_110561342_110561544 | 0.60 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1659 |
0.44 |
| chr1_239881158_239881438 | 0.57 |
ENSG00000233355 |
. |
1068 |
0.49 |
| chr14_22948566_22948717 | 0.57 |
ENSG00000251002 |
. |
1637 |
0.2 |
| chr7_50415328_50415483 | 0.57 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
48160 |
0.16 |
| chr17_29637369_29637894 | 0.57 |
EVI2B |
ecotropic viral integration site 2B |
3471 |
0.16 |
| chr1_198610920_198611071 | 0.56 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
2703 |
0.34 |
| chr14_38558504_38558837 | 0.56 |
CTD-2058B24.2 |
|
1693 |
0.48 |
| chr5_56125291_56125578 | 0.56 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
14033 |
0.16 |
| chr7_142429480_142429631 | 0.55 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
27764 |
0.17 |
| chr6_106970433_106970694 | 0.55 |
AIM1 |
absent in melanoma 1 |
10833 |
0.2 |
| chr4_40202894_40203061 | 0.54 |
RHOH |
ras homolog family member H |
1013 |
0.59 |
| chr17_38018967_38019134 | 0.54 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1329 |
0.36 |
| chr3_121373349_121373500 | 0.53 |
ENSG00000243544 |
. |
281 |
0.82 |
| chr1_111417492_111418003 | 0.52 |
CD53 |
CD53 molecule |
1971 |
0.34 |
| chr5_96294037_96294231 | 0.52 |
LNPEP |
leucyl/cystinyl aminopeptidase |
21 |
0.98 |
| chr2_199271076_199271227 | 0.51 |
ENSG00000252511 |
. |
160302 |
0.04 |
| chr1_101706338_101706489 | 0.51 |
RP4-575N6.5 |
|
2301 |
0.23 |
| chr14_88472278_88472651 | 0.50 |
GPR65 |
G protein-coupled receptor 65 |
996 |
0.51 |
| chr17_38716835_38717173 | 0.50 |
CCR7 |
chemokine (C-C motif) receptor 7 |
261 |
0.91 |
| chr10_65033773_65033924 | 0.49 |
JMJD1C |
jumonji domain containing 1C |
4866 |
0.31 |
| chr7_50345445_50345596 | 0.49 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1142 |
0.64 |
| chr1_169672728_169672969 | 0.48 |
SELL |
selectin L |
7991 |
0.2 |
| chr12_47611625_47611801 | 0.48 |
PCED1B |
PC-esterase domain containing 1B |
1332 |
0.49 |
| chr3_151916550_151916780 | 0.47 |
MBNL1 |
muscleblind-like splicing regulator 1 |
69164 |
0.11 |
| chr19_3136242_3137376 | 0.47 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
618 |
0.58 |
| chr3_151922152_151922513 | 0.47 |
MBNL1 |
muscleblind-like splicing regulator 1 |
63497 |
0.12 |
| chr10_8179903_8180054 | 0.47 |
GATA3 |
GATA binding protein 3 |
83209 |
0.11 |
| chr1_192546230_192546423 | 0.46 |
RGS1 |
regulator of G-protein signaling 1 |
1423 |
0.45 |
| chr2_225809390_225809894 | 0.46 |
DOCK10 |
dedicator of cytokinesis 10 |
2140 |
0.45 |
| chr10_62487545_62487752 | 0.46 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
5600 |
0.3 |
| chr11_118178183_118178334 | 0.46 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
2644 |
0.19 |
| chrX_51545134_51545285 | 0.46 |
MAGED1 |
melanoma antigen family D, 1 |
894 |
0.68 |
| chr3_85786117_85786268 | 0.45 |
CADM2 |
cell adhesion molecule 2 |
10560 |
0.3 |
| chr11_117873682_117873833 | 0.45 |
IL10RA |
interleukin 10 receptor, alpha |
16648 |
0.17 |
| chr13_46755525_46755771 | 0.45 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
811 |
0.58 |
| chr4_11473602_11473900 | 0.45 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
42362 |
0.18 |
| chr6_106968393_106968700 | 0.45 |
AIM1 |
absent in melanoma 1 |
8816 |
0.21 |
| chr13_24826594_24826745 | 0.45 |
SPATA13 |
spermatogenesis associated 13 |
813 |
0.56 |
| chr14_100532787_100533367 | 0.44 |
EVL |
Enah/Vasp-like |
303 |
0.88 |
| chr20_51618499_51618650 | 0.44 |
TSHZ2 |
teashirt zinc finger homeobox 2 |
29628 |
0.21 |
| chr2_158274562_158274945 | 0.44 |
CYTIP |
cytohesin 1 interacting protein |
21173 |
0.2 |
| chr1_102637571_102637722 | 0.44 |
OLFM3 |
olfactomedin 3 |
175060 |
0.04 |
| chr1_198624965_198625116 | 0.44 |
RP11-553K8.5 |
|
11150 |
0.23 |
| chr2_26045186_26045754 | 0.44 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
42292 |
0.17 |
| chr7_38389675_38390189 | 0.43 |
AMPH |
amphiphysin |
112781 |
0.07 |
| chr11_94960734_94960891 | 0.43 |
RP11-712B9.2 |
|
2538 |
0.31 |
| chr5_156616823_156617066 | 0.43 |
ITK |
IL2-inducible T-cell kinase |
9107 |
0.13 |
| chr4_143488164_143488362 | 0.43 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
6441 |
0.35 |
| chr14_22958589_22958740 | 0.43 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
2493 |
0.16 |
| chr2_158299271_158299480 | 0.43 |
CYTIP |
cytohesin 1 interacting protein |
1279 |
0.38 |
| chr5_35859051_35859218 | 0.42 |
IL7R |
interleukin 7 receptor |
2140 |
0.33 |
| chr5_156610422_156610951 | 0.42 |
ITK |
IL2-inducible T-cell kinase |
2849 |
0.18 |
| chrX_13680096_13680380 | 0.42 |
TCEANC |
transcription elongation factor A (SII) N-terminal and central domain containing |
6824 |
0.2 |
| chr6_35271134_35271290 | 0.42 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
5583 |
0.2 |
| chr2_68961577_68961872 | 0.42 |
ARHGAP25 |
Rho GTPase activating protein 25 |
189 |
0.96 |
| chr17_29644174_29644325 | 0.42 |
CTD-2370N5.3 |
|
1409 |
0.29 |
| chr1_167483926_167484083 | 0.42 |
CD247 |
CD247 molecule |
3771 |
0.25 |
| chr5_148727072_148727373 | 0.42 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
2157 |
0.2 |
| chr2_197033507_197033658 | 0.42 |
STK17B |
serine/threonine kinase 17b |
2142 |
0.32 |
| chr5_103563842_103563993 | 0.41 |
ENSG00000239808 |
. |
32610 |
0.25 |
| chr10_11213825_11214053 | 0.41 |
RP3-323N1.2 |
|
600 |
0.77 |
| chr1_32719471_32719691 | 0.41 |
LCK |
lymphocyte-specific protein tyrosine kinase |
2706 |
0.13 |
| chr12_19375312_19375463 | 0.41 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
14471 |
0.22 |
| chr2_99346083_99347252 | 0.40 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
44 |
0.98 |
| chr2_68978993_68979334 | 0.40 |
ARHGAP25 |
Rho GTPase activating protein 25 |
17149 |
0.23 |
| chr6_133084105_133084393 | 0.40 |
VNN2 |
vanin 2 |
321 |
0.83 |
| chr14_22771842_22772128 | 0.40 |
ENSG00000251002 |
. |
129734 |
0.04 |
| chr6_26361253_26361404 | 0.40 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
4059 |
0.11 |
| chr6_32556246_32556444 | 0.40 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
1280 |
0.36 |
| chr22_40858929_40859404 | 0.40 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
256 |
0.92 |
| chr20_39676850_39677439 | 0.39 |
TOP1 |
topoisomerase (DNA) I |
19686 |
0.2 |
| chr5_180243457_180243608 | 0.39 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
952 |
0.53 |
| chr15_38854702_38854853 | 0.39 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
2059 |
0.32 |
| chr13_49065972_49066123 | 0.39 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
41166 |
0.17 |
| chr18_51245241_51245392 | 0.39 |
RP11-671P2.1 |
|
150663 |
0.05 |
| chr9_88953252_88953465 | 0.39 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
4680 |
0.26 |
| chr14_42218476_42218627 | 0.39 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
49921 |
0.19 |
| chr1_174935570_174935876 | 0.39 |
RABGAP1L |
RAB GTPase activating protein 1-like |
1818 |
0.28 |
| chr2_158295494_158296053 | 0.39 |
CYTIP |
cytohesin 1 interacting protein |
153 |
0.96 |
| chr10_6619930_6620081 | 0.39 |
PRKCQ |
protein kinase C, theta |
2196 |
0.46 |
| chr21_30204766_30204917 | 0.39 |
N6AMT1 |
N-6 adenine-specific DNA methyltransferase 1 (putative) |
52852 |
0.14 |
| chr17_15586451_15586602 | 0.39 |
TRIM16 |
tripartite motif containing 16 |
333 |
0.83 |
| chr7_37381509_37381660 | 0.39 |
ELMO1 |
engulfment and cell motility 1 |
783 |
0.68 |
| chr1_198590020_198590171 | 0.39 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
17706 |
0.23 |
| chr21_36414032_36414216 | 0.39 |
RUNX1 |
runt-related transcription factor 1 |
7338 |
0.33 |
| chr13_41556163_41556400 | 0.38 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
137 |
0.96 |
| chr1_200859875_200861145 | 0.38 |
C1orf106 |
chromosome 1 open reading frame 106 |
41 |
0.97 |
| chrX_12990488_12990859 | 0.38 |
TMSB4X |
thymosin beta 4, X-linked |
2554 |
0.33 |
| chr6_90813429_90813580 | 0.38 |
ENSG00000222078 |
. |
102279 |
0.07 |
| chr4_83145308_83145459 | 0.38 |
ENSG00000202485 |
. |
49683 |
0.16 |
| chr6_45982477_45982717 | 0.38 |
CLIC5 |
chloride intracellular channel 5 |
968 |
0.6 |
| chr6_109223664_109223815 | 0.38 |
ARMC2-AS1 |
ARMC2 antisense RNA 1 |
21567 |
0.2 |
| chr1_229461570_229461782 | 0.38 |
RP4-803J11.2 |
|
6526 |
0.15 |
| chr6_144666173_144666513 | 0.38 |
UTRN |
utrophin |
1106 |
0.59 |
| chr1_9717913_9718080 | 0.38 |
C1orf200 |
chromosome 1 open reading frame 200 |
3352 |
0.21 |
| chr14_22887351_22887502 | 0.38 |
ENSG00000251002 |
. |
14293 |
0.12 |
| chr5_56473598_56474064 | 0.38 |
GPBP1 |
GC-rich promoter binding protein 1 |
2058 |
0.37 |
| chr1_117301810_117301961 | 0.37 |
CD2 |
CD2 molecule |
4796 |
0.25 |
| chr13_99992298_99992449 | 0.37 |
ENSG00000207719 |
. |
16012 |
0.19 |
| chr8_126960397_126960568 | 0.37 |
ENSG00000206695 |
. |
47287 |
0.2 |
| chr13_41551268_41551419 | 0.37 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
5075 |
0.22 |
| chr2_202124822_202124973 | 0.37 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
326 |
0.89 |
| chr16_27415626_27415898 | 0.37 |
IL21R |
interleukin 21 receptor |
1339 |
0.47 |
| chr10_17705373_17705524 | 0.37 |
ENSG00000251959 |
. |
15739 |
0.14 |
| chr9_128506323_128506474 | 0.37 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
3226 |
0.33 |
| chr1_161154335_161154531 | 0.36 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
7146 |
0.07 |
| chr2_106413120_106413271 | 0.36 |
NCK2 |
NCK adaptor protein 2 |
19820 |
0.25 |
| chr3_53077435_53077586 | 0.36 |
SFMBT1 |
Scm-like with four mbt domains 1 |
1771 |
0.34 |
| chr2_41127_41278 | 0.36 |
FAM110C |
family with sequence similarity 110, member C |
5183 |
0.33 |
| chr15_96422557_96422745 | 0.36 |
ENSG00000222076 |
. |
133618 |
0.06 |
| chr11_94886237_94886421 | 0.36 |
RP11-712B9.2 |
|
1419 |
0.51 |
| chr12_9760101_9760256 | 0.36 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
304 |
0.86 |
| chr10_28909540_28909691 | 0.36 |
ENSG00000252401 |
. |
31944 |
0.16 |
| chr2_7017776_7018587 | 0.36 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
385 |
0.85 |
| chr1_29253817_29254129 | 0.36 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12882 |
0.18 |
| chr14_22949288_22949494 | 0.36 |
ENSG00000251002 |
. |
887 |
0.35 |
| chr12_59460604_59460755 | 0.36 |
RP11-150C16.1 |
|
146259 |
0.04 |
| chr13_99966619_99966770 | 0.35 |
GPR183 |
G protein-coupled receptor 183 |
7035 |
0.21 |
| chr7_142423497_142423648 | 0.35 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
33747 |
0.16 |
| chr2_225734023_225734174 | 0.35 |
DOCK10 |
dedicator of cytokinesis 10 |
52729 |
0.18 |
| chr1_193506744_193506895 | 0.35 |
ENSG00000252241 |
. |
194255 |
0.03 |
| chr12_32120805_32121101 | 0.35 |
KIAA1551 |
KIAA1551 |
5530 |
0.26 |
| chrX_25812581_25812732 | 0.35 |
MAGEB18 |
melanoma antigen family B, 18 |
343804 |
0.01 |
| chr5_97126080_97126231 | 0.35 |
CTD-2215E18.2 |
|
588571 |
0.0 |
| chr10_73844664_73844815 | 0.35 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
3347 |
0.29 |
| chr6_76320868_76321019 | 0.35 |
SENP6 |
SUMO1/sentrin specific peptidase 6 |
9180 |
0.17 |
| chr21_19161404_19161758 | 0.35 |
AL109761.5 |
|
4224 |
0.27 |
| chr14_94437286_94437966 | 0.35 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
2803 |
0.22 |
| chr20_47436352_47436503 | 0.35 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
7993 |
0.28 |
| chr14_22636757_22636908 | 0.35 |
ENSG00000238634 |
. |
25945 |
0.24 |
| chr18_32180370_32180521 | 0.34 |
DTNA |
dystrobrevin, alpha |
7116 |
0.32 |
| chr3_63937239_63937390 | 0.34 |
ATXN7 |
ataxin 7 |
16106 |
0.16 |
| chr5_148726870_148727055 | 0.34 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
1897 |
0.22 |
| chr1_169672524_169672675 | 0.34 |
SELL |
selectin L |
8240 |
0.2 |
| chr1_158903283_158903483 | 0.34 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
2025 |
0.36 |
| chr7_115555128_115555279 | 0.34 |
TFEC |
transcription factor EC |
53101 |
0.16 |
| chr1_111745814_111746254 | 0.34 |
DENND2D |
DENN/MADD domain containing 2D |
997 |
0.41 |
| chr3_112692076_112692299 | 0.34 |
CD200R1 |
CD200 receptor 1 |
1572 |
0.33 |
| chr12_118796178_118796667 | 0.34 |
TAOK3 |
TAO kinase 3 |
488 |
0.85 |
| chr8_37755503_37755707 | 0.34 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
1367 |
0.34 |
| chr14_29078339_29078490 | 0.34 |
RP11-966I7.1 |
|
156055 |
0.04 |
| chr3_149419705_149419856 | 0.34 |
WWTR1 |
WW domain containing transcription regulator 1 |
1280 |
0.44 |
| chr3_18476194_18476345 | 0.34 |
SATB1 |
SATB homeobox 1 |
476 |
0.83 |
| chr3_43331814_43332301 | 0.34 |
SNRK |
SNF related kinase |
3979 |
0.23 |
| chr14_22972290_22972441 | 0.34 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
16194 |
0.09 |
| chr9_134151798_134152108 | 0.34 |
FAM78A |
family with sequence similarity 78, member A |
19 |
0.98 |
| chr11_314983_315695 | 0.34 |
IFITM1 |
interferon induced transmembrane protein 1 |
1486 |
0.17 |
| chr8_67053974_67054125 | 0.34 |
TRIM55 |
tripartite motif containing 55 |
14771 |
0.23 |
| chr14_102291094_102291439 | 0.33 |
CTD-2017C7.1 |
|
14602 |
0.14 |
| chr12_2410862_2411129 | 0.33 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
32053 |
0.21 |
| chr1_206718342_206718493 | 0.33 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
12076 |
0.16 |
| chr4_90220151_90220996 | 0.33 |
GPRIN3 |
GPRIN family member 3 |
8588 |
0.31 |
| chr1_215313373_215313524 | 0.33 |
KCNK2 |
potassium channel, subfamily K, member 2 |
56588 |
0.18 |
| chr4_143488982_143489737 | 0.32 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
7537 |
0.34 |
| chr11_46581548_46581763 | 0.32 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
31259 |
0.15 |
| chr12_113344776_113345099 | 0.32 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
80 |
0.97 |
| chr14_32672059_32672251 | 0.32 |
ENSG00000202337 |
. |
320 |
0.8 |
| chr3_195265081_195265237 | 0.32 |
PPP1R2 |
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
5017 |
0.17 |
| chr5_162935820_162936429 | 0.32 |
MAT2B |
methionine adenosyltransferase II, beta |
3504 |
0.21 |
| chr11_39392973_39393210 | 0.32 |
ENSG00000201591 |
. |
110434 |
0.08 |
| chr6_25040660_25040811 | 0.32 |
RP3-425P12.5 |
|
1332 |
0.34 |
| chr21_43825650_43825846 | 0.32 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
1722 |
0.26 |
| chr11_116802141_116802415 | 0.32 |
SIK3 |
SIK family kinase 3 |
25482 |
0.14 |
| chr14_91877157_91877308 | 0.32 |
CCDC88C |
coiled-coil domain containing 88C |
6458 |
0.25 |
| chr2_204803308_204803520 | 0.32 |
ICOS |
inducible T-cell co-stimulator |
1911 |
0.46 |
| chr12_98912505_98912656 | 0.32 |
TMPO-AS1 |
TMPO antisense RNA 1 |
2380 |
0.2 |
| chr1_111421280_111421431 | 0.32 |
CD53 |
CD53 molecule |
5579 |
0.21 |
| chr1_114413652_114413958 | 0.32 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
515 |
0.69 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.2 | 0.3 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.7 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.1 | 0.4 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 1.2 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.4 | GO:0002517 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.1 | 0.8 | GO:0002836 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.1 | 0.5 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.1 | 0.3 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.1 | 0.3 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.2 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 0.2 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.6 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.1 | 0.2 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.3 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.2 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.3 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.4 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.1 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.6 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.0 | GO:0070897 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.3 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.0 | GO:1903306 | negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.3 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.2 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.8 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.8 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.0 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.6 | GO:0001892 | embryonic placenta development(GO:0001892) |
| 0.0 | 0.3 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:1904590 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein localization to nucleus(GO:1900181) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0014824 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.0 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.0 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.0 | GO:0048617 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0035929 | steroid hormone secretion(GO:0035929) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.5 | GO:0097194 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.0 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.0 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.7 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 1.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 1.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.5 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 2.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.0 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.0 | REACTOME S PHASE | Genes involved in S Phase |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |