Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
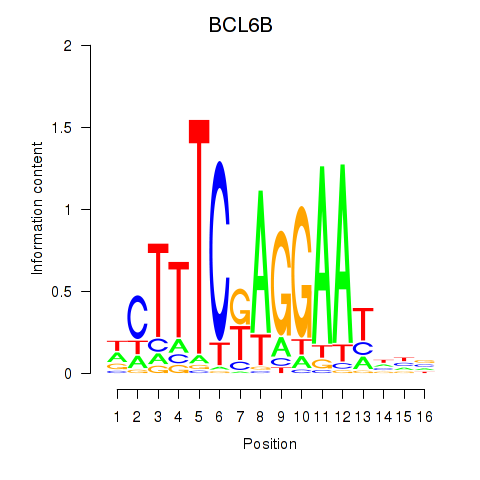
Results for BCL6B
Z-value: 1.58
Transcription factors associated with BCL6B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BCL6B
|
ENSG00000161940.6 | BCL6B transcription repressor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_6927262_6927413 | BCL6B | 541 | 0.501489 | -0.65 | 6.0e-02 | Click! |
| chr17_6926858_6927139 | BCL6B | 202 | 0.825004 | -0.63 | 7.0e-02 | Click! |
| chr17_6926552_6926816 | BCL6B | 112 | 0.893632 | -0.55 | 1.2e-01 | Click! |
| chr17_6927436_6927587 | BCL6B | 715 | 0.383842 | -0.38 | 3.2e-01 | Click! |
| chr17_6932977_6933147 | BCL6B | 3242 | 0.089383 | -0.33 | 3.9e-01 | Click! |
Activity of the BCL6B motif across conditions
Conditions sorted by the z-value of the BCL6B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
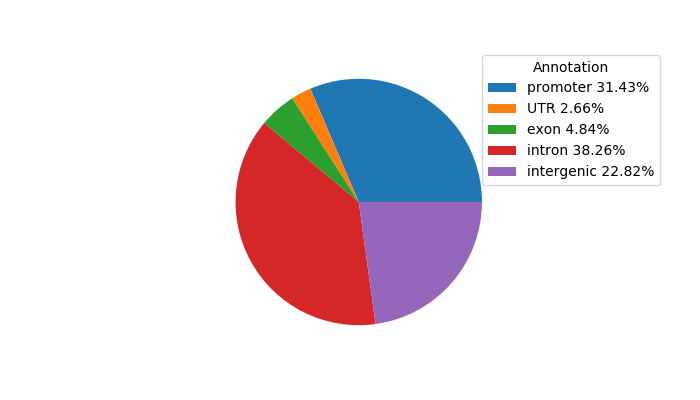
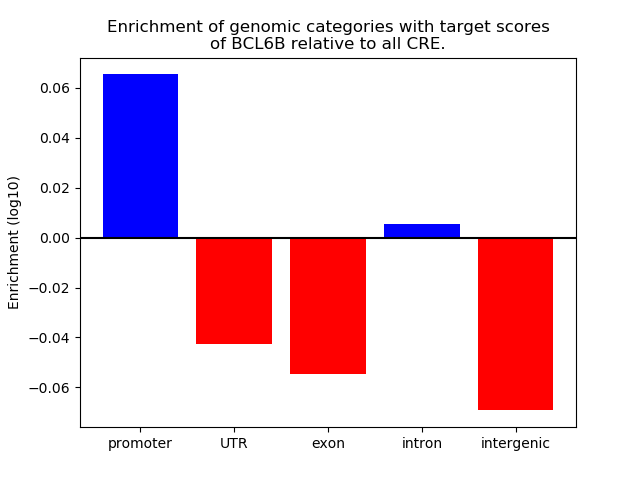
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_55513844_55514378 | 0.58 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
201 |
0.95 |
| chr9_117880836_117880987 | 0.56 |
TNC |
tenascin C |
375 |
0.9 |
| chr2_238321930_238322164 | 0.55 |
COL6A3 |
collagen, type VI, alpha 3 |
744 |
0.69 |
| chr5_33891044_33891453 | 0.54 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
798 |
0.63 |
| chr6_27721768_27721989 | 0.43 |
HIST1H2BL |
histone cluster 1, H2bl |
53831 |
0.06 |
| chr19_1591177_1591368 | 0.42 |
MBD3 |
methyl-CpG binding domain protein 3 |
1380 |
0.23 |
| chr13_21475841_21475992 | 0.41 |
XPO4 |
exportin 4 |
997 |
0.6 |
| chr4_99578892_99579431 | 0.41 |
TSPAN5 |
tetraspanin 5 |
318 |
0.81 |
| chr4_81198150_81198317 | 0.41 |
FGF5 |
fibroblast growth factor 5 |
10440 |
0.25 |
| chr11_62621592_62621969 | 0.41 |
ENSG00000255717 |
. |
23 |
0.93 |
| chr7_19155354_19155672 | 0.40 |
TWIST1 |
twist family bHLH transcription factor 1 |
1782 |
0.28 |
| chr11_14379258_14380093 | 0.40 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
356 |
0.92 |
| chr6_130088124_130088932 | 0.40 |
ENSG00000211996 |
. |
19646 |
0.24 |
| chr2_1748699_1748904 | 0.40 |
PXDN |
peroxidasin homolog (Drosophila) |
523 |
0.86 |
| chr11_62320728_62321001 | 0.39 |
AHNAK |
AHNAK nucleoprotein |
2843 |
0.12 |
| chr1_147013251_147013702 | 0.39 |
BCL9 |
B-cell CLL/lymphoma 9 |
294 |
0.94 |
| chr2_217365937_217366088 | 0.38 |
RPL37A |
ribosomal protein L37a |
2149 |
0.24 |
| chr18_67957874_67958133 | 0.38 |
SOCS6 |
suppressor of cytokine signaling 6 |
1866 |
0.41 |
| chr20_2634564_2634928 | 0.37 |
ENSG00000221116 |
. |
112 |
0.85 |
| chr4_8594931_8595082 | 0.37 |
CPZ |
carboxypeptidase Z |
523 |
0.82 |
| chr22_46478725_46478917 | 0.36 |
FLJ27365 |
hsa-mir-4763 |
2629 |
0.15 |
| chr22_39713562_39713713 | 0.36 |
RPL3 |
ribosomal protein L3 |
982 |
0.32 |
| chr3_123602332_123603013 | 0.36 |
MYLK |
myosin light chain kinase |
477 |
0.85 |
| chr12_113859774_113860410 | 0.35 |
SDSL |
serine dehydratase-like |
50 |
0.97 |
| chr2_216299709_216300678 | 0.35 |
FN1 |
fibronectin 1 |
597 |
0.55 |
| chr12_42877884_42878224 | 0.35 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
80 |
0.97 |
| chr20_43975844_43976401 | 0.34 |
SDC4 |
syndecan 4 |
942 |
0.43 |
| chr1_213123311_213123462 | 0.34 |
VASH2 |
vasohibin 2 |
476 |
0.85 |
| chr11_844288_844547 | 0.33 |
TSPAN4 |
tetraspanin 4 |
29 |
0.92 |
| chr4_143325352_143326244 | 0.33 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
26614 |
0.28 |
| chr2_112895388_112895739 | 0.33 |
FBLN7 |
fibulin 7 |
399 |
0.89 |
| chr4_86435677_86435949 | 0.33 |
ARHGAP24 |
Rho GTPase activating protein 24 |
39422 |
0.22 |
| chr5_136833711_136834156 | 0.33 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
314 |
0.93 |
| chr7_129913504_129913799 | 0.32 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
6984 |
0.15 |
| chr11_120208214_120208431 | 0.32 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
376 |
0.86 |
| chr1_156675058_156675325 | 0.32 |
CRABP2 |
cellular retinoic acid binding protein 2 |
202 |
0.87 |
| chrX_11446077_11446283 | 0.31 |
ARHGAP6 |
Rho GTPase activating protein 6 |
287 |
0.95 |
| chr1_227504067_227504291 | 0.31 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
704 |
0.8 |
| chr11_58875134_58875345 | 0.31 |
FAM111B |
family with sequence similarity 111, member B |
529 |
0.77 |
| chr7_44922298_44922675 | 0.31 |
ENSG00000264326 |
. |
1087 |
0.34 |
| chr6_128839981_128840137 | 0.31 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
1444 |
0.38 |
| chrX_2610555_2610727 | 0.30 |
CD99 |
CD99 molecule |
1100 |
0.58 |
| chr4_152021608_152021943 | 0.30 |
RPS3A |
ribosomal protein S3A |
750 |
0.52 |
| chr10_122216113_122216362 | 0.30 |
PPAPDC1A |
phosphatidic acid phosphatase type 2 domain containing 1A |
229 |
0.95 |
| chr20_17523751_17523902 | 0.30 |
BFSP1 |
beaded filament structural protein 1, filensin |
11812 |
0.2 |
| chr6_10695950_10696101 | 0.30 |
C6orf52 |
chromosome 6 open reading frame 52 |
995 |
0.33 |
| chr11_65324607_65325239 | 0.30 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
313 |
0.76 |
| chr11_27956142_27956384 | 0.30 |
RP11-587D21.4 |
|
57068 |
0.15 |
| chrY_2560555_2560707 | 0.30 |
ENSG00000251841 |
. |
92159 |
0.09 |
| chr11_93863372_93863641 | 0.30 |
PANX1 |
pannexin 1 |
1411 |
0.53 |
| chr4_99069479_99069630 | 0.29 |
STPG2 |
sperm-tail PG-rich repeat containing 2 |
5163 |
0.26 |
| chr3_119896297_119896512 | 0.29 |
ENSG00000206721 |
. |
32763 |
0.16 |
| chr12_7154780_7154931 | 0.29 |
C1S |
complement component 1, s subcomponent |
12964 |
0.1 |
| chr2_238341641_238342175 | 0.29 |
AC112721.1 |
Uncharacterized protein |
7989 |
0.2 |
| chr3_191048468_191048928 | 0.29 |
UTS2B |
urotensin 2B |
373 |
0.85 |
| chr3_77089535_77089704 | 0.29 |
ROBO2 |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
262 |
0.95 |
| chr3_185960084_185960372 | 0.29 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
38275 |
0.19 |
| chr1_172109378_172109611 | 0.29 |
ENSG00000207949 |
. |
1447 |
0.39 |
| chr14_34929919_34930181 | 0.29 |
SPTSSA |
serine palmitoyltransferase, small subunit A |
1512 |
0.47 |
| chr10_24756203_24756381 | 0.29 |
KIAA1217 |
KIAA1217 |
832 |
0.69 |
| chr15_75930684_75930835 | 0.29 |
IMP3 |
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
1793 |
0.19 |
| chr1_190447721_190448034 | 0.28 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
1118 |
0.69 |
| chr4_7878163_7878536 | 0.28 |
AFAP1 |
actin filament associated protein 1 |
4309 |
0.28 |
| chr1_74663321_74663472 | 0.28 |
LRRIQ3 |
leucine-rich repeats and IQ motif containing 3 |
435 |
0.3 |
| chr16_85269227_85269622 | 0.28 |
CTC-786C10.1 |
|
64542 |
0.11 |
| chr11_123065103_123065500 | 0.28 |
CLMP |
CXADR-like membrane protein |
688 |
0.71 |
| chr5_82768204_82768925 | 0.28 |
VCAN |
versican |
820 |
0.75 |
| chr12_13349672_13350378 | 0.28 |
EMP1 |
epithelial membrane protein 1 |
305 |
0.93 |
| chr8_117883179_117883330 | 0.28 |
RAD21 |
RAD21 homolog (S. pombe) |
3345 |
0.19 |
| chr1_146715427_146715640 | 0.28 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
1177 |
0.49 |
| chr19_42581255_42581466 | 0.28 |
ZNF574 |
zinc finger protein 574 |
1042 |
0.35 |
| chr7_94279563_94279714 | 0.28 |
SGCE |
sarcoglycan, epsilon |
5772 |
0.22 |
| chr11_122754419_122754570 | 0.27 |
C11orf63 |
chromosome 11 open reading frame 63 |
1021 |
0.55 |
| chr12_110720958_110721647 | 0.27 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
1816 |
0.41 |
| chr14_52121858_52122101 | 0.27 |
FRMD6 |
FERM domain containing 6 |
3281 |
0.25 |
| chr8_125382455_125382617 | 0.27 |
TMEM65 |
transmembrane protein 65 |
2397 |
0.35 |
| chr6_158405199_158405350 | 0.27 |
SYNJ2 |
synaptojanin 2 |
2204 |
0.3 |
| chr9_21357722_21357949 | 0.27 |
IFNA6 |
interferon, alpha 6 |
6458 |
0.13 |
| chr12_122983015_122983166 | 0.26 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
782 |
0.59 |
| chr2_203132095_203132497 | 0.26 |
NOP58 |
NOP58 ribonucleoprotein |
1857 |
0.21 |
| chr1_193029578_193029796 | 0.26 |
UCHL5 |
ubiquitin carboxyl-terminal hydrolase L5 |
450 |
0.47 |
| chr4_113739998_113740296 | 0.26 |
ANK2 |
ankyrin 2, neuronal |
882 |
0.65 |
| chr11_94513607_94513844 | 0.26 |
AMOTL1 |
angiomotin like 1 |
12188 |
0.23 |
| chrX_128655749_128656001 | 0.26 |
SMARCA1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
1585 |
0.44 |
| chr7_148724335_148724802 | 0.26 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
992 |
0.45 |
| chr18_12310772_12310961 | 0.26 |
TUBB6 |
tubulin, beta 6 class V |
100 |
0.97 |
| chr11_93582978_93583471 | 0.25 |
VSTM5 |
V-set and transmembrane domain containing 5 |
473 |
0.74 |
| chr17_41984921_41985550 | 0.25 |
MPP2 |
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
108 |
0.94 |
| chr17_66950763_66951090 | 0.25 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
456 |
0.87 |
| chr6_80852871_80853089 | 0.25 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
36597 |
0.23 |
| chr10_112258170_112258629 | 0.25 |
DUSP5 |
dual specificity phosphatase 5 |
803 |
0.63 |
| chr13_108518334_108518485 | 0.25 |
FAM155A |
family with sequence similarity 155, member A |
674 |
0.78 |
| chr17_68133789_68133994 | 0.25 |
AC002539.1 |
|
8891 |
0.19 |
| chr17_79069332_79069483 | 0.25 |
BAIAP2 |
BAI1-associated protein 2 |
1972 |
0.21 |
| chr6_28366113_28366264 | 0.25 |
ZSCAN12 |
zinc finger and SCAN domain containing 12 |
1320 |
0.37 |
| chr16_3449281_3449432 | 0.24 |
ZSCAN32 |
zinc finger and SCAN domain containing 32 |
1608 |
0.22 |
| chr10_96121255_96121406 | 0.24 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
381 |
0.87 |
| chr8_23837042_23837198 | 0.24 |
ENSG00000207201 |
. |
97551 |
0.07 |
| chr4_109967516_109967667 | 0.24 |
COL25A1 |
collagen, type XXV, alpha 1 |
222216 |
0.02 |
| chr6_44900994_44901181 | 0.24 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
22160 |
0.28 |
| chrX_34673939_34674563 | 0.24 |
TMEM47 |
transmembrane protein 47 |
1154 |
0.68 |
| chr12_86229562_86230264 | 0.24 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
435 |
0.89 |
| chr12_97379793_97379962 | 0.24 |
ENSG00000202368 |
. |
6470 |
0.31 |
| chr6_28953244_28953516 | 0.24 |
HCG15 |
HLA complex group 15 (non-protein coding) |
600 |
0.54 |
| chr1_11867192_11867412 | 0.24 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
325 |
0.73 |
| chr20_52829309_52829550 | 0.24 |
PFDN4 |
prefoldin subunit 4 |
5043 |
0.28 |
| chrX_57148342_57148596 | 0.24 |
RP3-323P24.3 |
|
374 |
0.57 |
| chr6_86159887_86160764 | 0.24 |
NT5E |
5'-nucleotidase, ecto (CD73) |
498 |
0.87 |
| chr8_23837345_23837496 | 0.24 |
ENSG00000207201 |
. |
97251 |
0.08 |
| chr2_12928575_12928807 | 0.24 |
ENSG00000264370 |
. |
51198 |
0.17 |
| chr12_54394768_54394919 | 0.24 |
HOXC9 |
homeobox C9 |
940 |
0.22 |
| chr11_133939751_133939941 | 0.24 |
JAM3 |
junctional adhesion molecule 3 |
880 |
0.67 |
| chr16_84571908_84572059 | 0.24 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
15656 |
0.17 |
| chr1_43637158_43637389 | 0.24 |
WDR65 |
WD repeat domain 65 |
547 |
0.48 |
| chr16_86599089_86599240 | 0.24 |
FOXC2 |
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
1693 |
0.28 |
| chr6_116718983_116719178 | 0.24 |
DSE |
dermatan sulfate epimerase |
26970 |
0.15 |
| chr4_54967212_54967377 | 0.24 |
GSX2 |
GS homeobox 2 |
954 |
0.49 |
| chr1_168329622_168329773 | 0.24 |
ENSG00000207974 |
. |
15065 |
0.23 |
| chr5_14665884_14666035 | 0.24 |
FAM105B |
family with sequence similarity 105, member B |
1102 |
0.41 |
| chr9_124414074_124414751 | 0.23 |
DAB2IP |
DAB2 interacting protein |
539 |
0.84 |
| chr3_157156592_157156743 | 0.23 |
PTX3 |
pentraxin 3, long |
2089 |
0.41 |
| chr6_11095182_11095333 | 0.23 |
SMIM13 |
small integral membrane protein 13 |
991 |
0.49 |
| chr7_44837548_44837765 | 0.23 |
PPIA |
peptidylprolyl isomerase A (cyclophilin A) |
1376 |
0.35 |
| chr8_32441306_32441509 | 0.23 |
NRG1 |
neuregulin 1 |
21759 |
0.28 |
| chr19_38145180_38145331 | 0.23 |
ZFP30 |
ZFP30 zinc finger protein |
375 |
0.84 |
| chr7_45243750_45244089 | 0.23 |
RAMP3 |
receptor (G protein-coupled) activity modifying protein 3 |
46491 |
0.13 |
| chr13_49687457_49688000 | 0.23 |
FNDC3A |
fibronectin type III domain containing 3A |
3279 |
0.28 |
| chr4_123379678_123379913 | 0.23 |
IL2 |
interleukin 2 |
1915 |
0.44 |
| chr1_163038601_163039117 | 0.23 |
RGS4 |
regulator of G-protein signaling 4 |
76 |
0.98 |
| chr3_160940495_160941006 | 0.23 |
NMD3 |
NMD3 ribosome export adaptor |
1403 |
0.58 |
| chr19_3671861_3672012 | 0.23 |
AC004637.1 |
|
644 |
0.58 |
| chr5_140480603_140480754 | 0.23 |
PCDHB3 |
protocadherin beta 3 |
444 |
0.6 |
| chr1_68178990_68179141 | 0.23 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
28181 |
0.18 |
| chr12_94135787_94136106 | 0.23 |
RP11-887P2.5 |
|
4347 |
0.27 |
| chr6_153413419_153413831 | 0.23 |
RGS17 |
regulator of G-protein signaling 17 |
38759 |
0.14 |
| chr6_112102070_112102221 | 0.22 |
FYN |
FYN oncogene related to SRC, FGR, YES |
138 |
0.98 |
| chr1_33206819_33207020 | 0.22 |
KIAA1522 |
KIAA1522 |
567 |
0.7 |
| chr5_322963_323164 | 0.22 |
AHRR |
aryl-hydrocarbon receptor repressor |
308 |
0.89 |
| chr4_153303627_153303899 | 0.22 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
96 |
0.98 |
| chr5_180685889_180686229 | 0.22 |
TRIM52 |
tripartite motif containing 52 |
2060 |
0.13 |
| chr6_35458145_35458450 | 0.22 |
TEAD3 |
TEA domain family member 3 |
6430 |
0.17 |
| chr20_30305454_30306019 | 0.22 |
AL160175.1 |
|
3265 |
0.17 |
| chr6_100381252_100381443 | 0.22 |
MCHR2-AS1 |
MCHR2 antisense RNA 1 |
60473 |
0.14 |
| chr10_33964074_33964254 | 0.22 |
NRP1 |
neuropilin 1 |
338974 |
0.01 |
| chr7_44921425_44922084 | 0.22 |
ENSG00000264326 |
. |
355 |
0.79 |
| chr11_114167265_114167463 | 0.22 |
NNMT |
nicotinamide N-methyltransferase |
721 |
0.72 |
| chr9_21803022_21803324 | 0.22 |
MTAP |
methylthioadenosine phosphorylase |
525 |
0.78 |
| chr3_71352476_71352627 | 0.22 |
FOXP1 |
forkhead box P1 |
1360 |
0.52 |
| chr8_29206404_29207399 | 0.22 |
DUSP4 |
dual specificity phosphatase 4 |
579 |
0.71 |
| chr2_65527534_65527956 | 0.22 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
34162 |
0.18 |
| chr1_182991327_182991478 | 0.22 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
1193 |
0.49 |
| chr12_93964096_93964684 | 0.22 |
SOCS2 |
suppressor of cytokine signaling 2 |
215 |
0.85 |
| chr12_131358148_131358299 | 0.22 |
RAN |
RAN, member RAS oncogene family |
1582 |
0.33 |
| chr2_36585229_36585663 | 0.22 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
1832 |
0.5 |
| chr7_15724850_15725001 | 0.22 |
MEOX2 |
mesenchyme homeobox 2 |
1512 |
0.49 |
| chr21_17567832_17567983 | 0.22 |
ENSG00000201025 |
. |
89182 |
0.1 |
| chrX_11682905_11683408 | 0.21 |
ARHGAP6 |
Rho GTPase activating protein 6 |
208 |
0.97 |
| chr22_38864095_38864404 | 0.21 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
166 |
0.93 |
| chrX_99899287_99899446 | 0.21 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
151 |
0.95 |
| chr10_99407246_99407471 | 0.21 |
PI4K2A |
phosphatidylinositol 4-kinase type 2 alpha |
6915 |
0.14 |
| chrX_56755732_56756297 | 0.21 |
ENSG00000204272 |
. |
322 |
0.94 |
| chr6_136823903_136824054 | 0.21 |
MAP7 |
microtubule-associated protein 7 |
23143 |
0.18 |
| chr9_16267939_16268151 | 0.21 |
C9orf92 |
chromosome 9 open reading frame 92 |
8266 |
0.32 |
| chr15_36699845_36700050 | 0.21 |
C15orf41 |
chromosome 15 open reading frame 41 |
171865 |
0.04 |
| chr1_144929565_144929716 | 0.21 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
2508 |
0.23 |
| chr17_17863110_17863392 | 0.21 |
TOM1L2 |
target of myb1-like 2 (chicken) |
12460 |
0.15 |
| chr17_41812174_41812325 | 0.21 |
ENSG00000215964 |
. |
13319 |
0.13 |
| chr20_25229665_25230039 | 0.21 |
PYGB |
phosphorylase, glycogen; brain |
1147 |
0.46 |
| chr5_138944731_138945044 | 0.21 |
UBE2D2 |
ubiquitin-conjugating enzyme E2D 2 |
3273 |
0.22 |
| chr6_56820874_56821079 | 0.21 |
BEND6 |
BEN domain containing 6 |
806 |
0.54 |
| chr7_135432429_135432583 | 0.21 |
FAM180A |
family with sequence similarity 180, member A |
954 |
0.53 |
| chr2_85768416_85768567 | 0.21 |
MAT2A |
methionine adenosyltransferase II, alpha |
1726 |
0.2 |
| chr15_60060576_60060814 | 0.21 |
BNIP2 |
BCL2/adenovirus E1B 19kDa interacting protein 2 |
78962 |
0.08 |
| chr1_92495050_92495201 | 0.21 |
EPHX4 |
epoxide hydrolase 4 |
414 |
0.87 |
| chr7_121784101_121784445 | 0.21 |
AASS |
aminoadipate-semialdehyde synthase |
61 |
0.98 |
| chr2_102930347_102930498 | 0.21 |
IL1RL1 |
interleukin 1 receptor-like 1 |
2394 |
0.26 |
| chr5_42423266_42423460 | 0.21 |
GHR |
growth hormone receptor |
516 |
0.88 |
| chr4_152022521_152022672 | 0.21 |
ENSG00000201264 |
. |
613 |
0.6 |
| chr11_120080847_120081389 | 0.21 |
OAF |
OAF homolog (Drosophila) |
357 |
0.86 |
| chr10_69645958_69646116 | 0.21 |
SIRT1 |
sirtuin 1 |
1098 |
0.54 |
| chrX_70505081_70505341 | 0.21 |
NONO |
non-POU domain containing, octamer-binding |
1167 |
0.43 |
| chr12_22200754_22200905 | 0.21 |
CMAS |
cytidine monophosphate N-acetylneuraminic acid synthetase |
1721 |
0.49 |
| chr3_16217443_16217716 | 0.20 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
1363 |
0.53 |
| chr4_114899439_114899761 | 0.20 |
ARSJ |
arylsulfatase family, member J |
552 |
0.84 |
| chr3_156269176_156269606 | 0.20 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
2523 |
0.33 |
| chr11_133789632_133789917 | 0.20 |
ENSG00000264919 |
. |
21298 |
0.17 |
| chr18_21974914_21975156 | 0.20 |
OSBPL1A |
oxysterol binding protein-like 1A |
2717 |
0.21 |
| chr1_86048499_86048650 | 0.20 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
2130 |
0.3 |
| chr7_86783544_86783895 | 0.20 |
DMTF1 |
cyclin D binding myb-like transcription factor 1 |
1772 |
0.31 |
| chr17_33409582_33409733 | 0.20 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
6197 |
0.11 |
| chr18_73496031_73496230 | 0.20 |
SMIM21 |
small integral membrane protein 21 |
356472 |
0.01 |
| chr12_125815735_125816035 | 0.20 |
TMEM132B |
transmembrane protein 132B |
4723 |
0.36 |
| chr15_96878987_96879612 | 0.20 |
ENSG00000222651 |
. |
2809 |
0.21 |
| chr5_139691412_139691728 | 0.20 |
PFDN1 |
prefoldin subunit 1 |
8864 |
0.14 |
| chr16_65151818_65151997 | 0.20 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
3926 |
0.39 |
| chr3_131220949_131221185 | 0.20 |
MRPL3 |
mitochondrial ribosomal protein L3 |
714 |
0.66 |
| chr1_113499835_113500197 | 0.20 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
381 |
0.75 |
| chr2_12861098_12861380 | 0.20 |
TRIB2 |
tribbles pseudokinase 2 |
2875 |
0.34 |
| chr7_113996511_113996662 | 0.20 |
FOXP2 |
forkhead box P2 |
57743 |
0.15 |
| chr12_64423145_64423296 | 0.20 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
46915 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.4 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.3 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.6 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 0.2 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.1 | GO:0072143 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.1 | 0.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.1 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0072239 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.0 | 0.3 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.2 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.3 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 1.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0060391 | regulation of SMAD protein import into nucleus(GO:0060390) positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:2001234 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.1 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.2 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.2 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.0 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.4 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.2 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0010042 | response to manganese ion(GO:0010042) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0070875 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0031645 | negative regulation of neurological system process(GO:0031645) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0097480 | synaptic vesicle transport(GO:0048489) synaptic vesicle localization(GO:0097479) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.3 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.0 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.0 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.8 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0010092 | specification of organ identity(GO:0010092) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.2 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0090114 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.6 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.2 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.0 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.1 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.0 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0043552 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 | 0.0 | GO:0035082 | axoneme assembly(GO:0035082) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.1 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.4 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 3.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.0 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 2.1 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.3 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.5 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.1 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.0 | 0.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.4 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.0 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |