Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
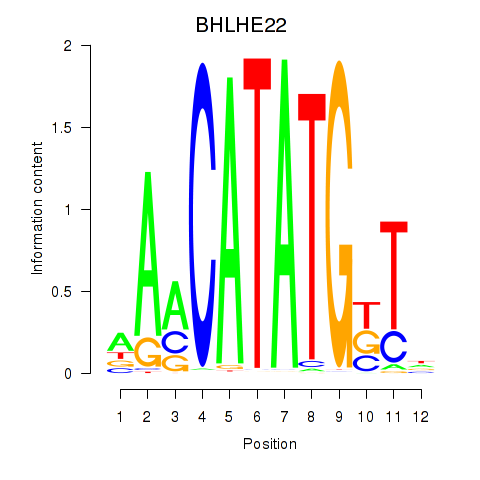
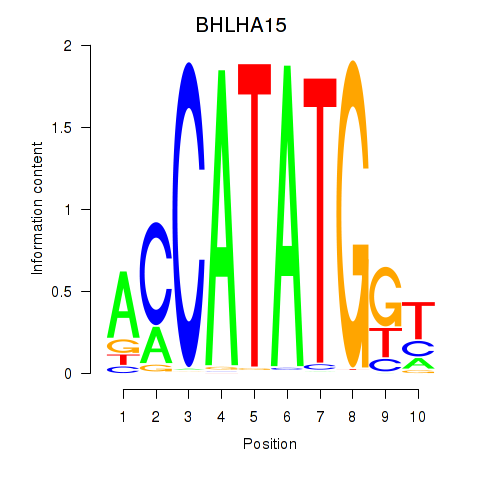
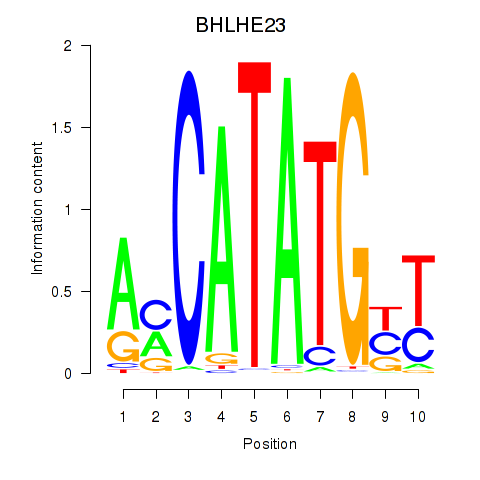
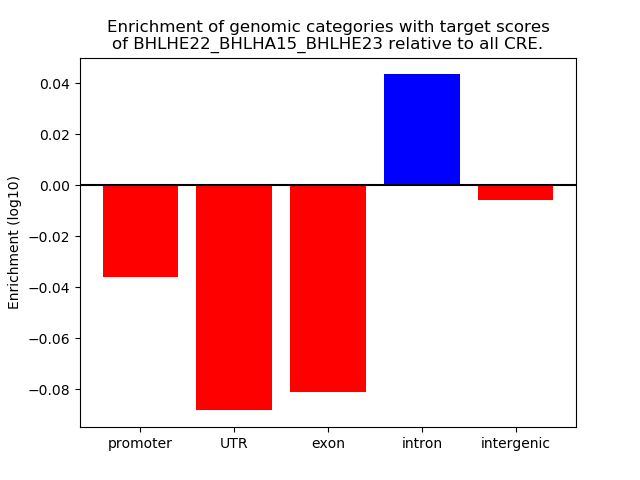
Results for BHLHE22_BHLHA15_BHLHE23
Z-value: 0.61
Transcription factors associated with BHLHE22_BHLHA15_BHLHE23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE22
|
ENSG00000180828.1 | basic helix-loop-helix family member e22 |
|
BHLHA15
|
ENSG00000180535.3 | basic helix-loop-helix family member a15 |
|
BHLHE23
|
ENSG00000125533.4 | basic helix-loop-helix family member e23 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_97793907_97794058 | BHLHA15 | 46757 | 0.129530 | 0.67 | 4.7e-02 | Click! |
| chr7_97840727_97841036 | BHLHA15 | 142 | 0.964058 | 0.65 | 5.6e-02 | Click! |
| chr7_97841044_97841241 | BHLHA15 | 403 | 0.864408 | 0.51 | 1.6e-01 | Click! |
| chr7_97794262_97794413 | BHLHA15 | 46402 | 0.130377 | 0.40 | 2.8e-01 | Click! |
| chr7_97841269_97841420 | BHLHA15 | 222 | 0.941200 | -0.39 | 2.9e-01 | Click! |
| chr8_65492553_65493166 | BHLHE22 | 45 | 0.973369 | 0.58 | 1.0e-01 | Click! |
| chr8_65492035_65492186 | BHLHE22 | 704 | 0.701291 | -0.42 | 2.6e-01 | Click! |
| chr8_65493405_65493750 | BHLHE22 | 763 | 0.482073 | 0.41 | 2.7e-01 | Click! |
| chr20_61640452_61640603 | BHLHE23 | 2140 | 0.299363 | 0.60 | 8.8e-02 | Click! |
| chr20_61613239_61613390 | BHLHE23 | 25073 | 0.130156 | 0.50 | 1.7e-01 | Click! |
| chr20_61640182_61640333 | BHLHE23 | 1870 | 0.329630 | 0.50 | 1.7e-01 | Click! |
| chr20_61656853_61657255 | BHLHE23 | 18667 | 0.168471 | -0.34 | 3.6e-01 | Click! |
| chr20_61641850_61642001 | BHLHE23 | 3538 | 0.227710 | 0.20 | 6.1e-01 | Click! |
Activity of the BHLHE22_BHLHA15_BHLHE23 motif across conditions
Conditions sorted by the z-value of the BHLHE22_BHLHA15_BHLHE23 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
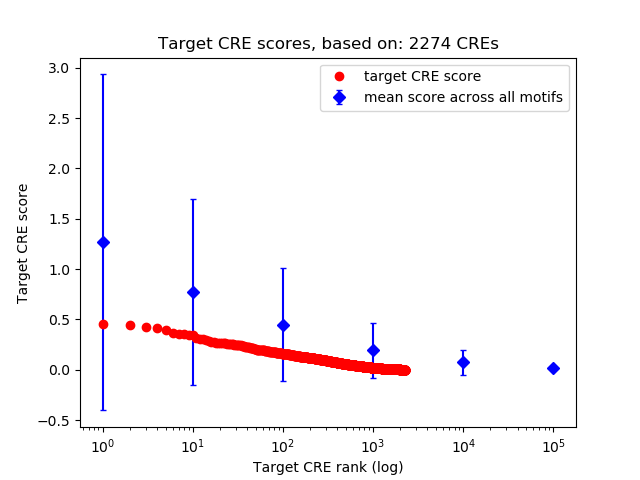
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_230307234_230307385 | 0.46 |
RP5-956O18.2 |
|
96920 |
0.07 |
| chr10_63979683_63979834 | 0.44 |
RTKN2 |
rhotekin 2 |
16264 |
0.26 |
| chr10_99302459_99302711 | 0.43 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
29613 |
0.09 |
| chr1_185283352_185283831 | 0.41 |
IVNS1ABP |
influenza virus NS1A binding protein |
2870 |
0.28 |
| chr18_56244732_56244930 | 0.40 |
RP11-126O1.2 |
|
20529 |
0.14 |
| chr8_122738565_122738716 | 0.37 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
84964 |
0.1 |
| chr7_106299749_106299900 | 0.36 |
CCDC71L |
coiled-coil domain containing 71-like |
1618 |
0.46 |
| chr2_200490274_200490425 | 0.35 |
SATB2 |
SATB homeobox 2 |
154360 |
0.04 |
| chr10_89307360_89307659 | 0.35 |
ENSG00000222192 |
. |
15434 |
0.21 |
| chr3_192675469_192675839 | 0.34 |
MB21D2 |
Mab-21 domain containing 2 |
39704 |
0.21 |
| chr7_97982123_97982542 | 0.31 |
RP11-307C18.1 |
|
30167 |
0.16 |
| chr10_26775632_26775856 | 0.31 |
ENSG00000199733 |
. |
22774 |
0.23 |
| chr6_154780043_154780194 | 0.30 |
CNKSR3 |
CNKSR family member 3 |
19507 |
0.27 |
| chr5_95295455_95295746 | 0.30 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
1952 |
0.29 |
| chr2_175264635_175264786 | 0.29 |
SCRN3 |
secernin 3 |
1787 |
0.32 |
| chr8_107621172_107621323 | 0.28 |
OXR1 |
oxidation resistance 1 |
9107 |
0.27 |
| chr5_3311555_3311706 | 0.28 |
IRX1 |
iroquois homeobox 1 |
284538 |
0.01 |
| chr5_14184300_14184719 | 0.27 |
TRIO |
trio Rho guanine nucleotide exchange factor |
602 |
0.86 |
| chr10_123458202_123458353 | 0.26 |
RP11-78A18.2 |
|
37541 |
0.2 |
| chr5_73516595_73516746 | 0.26 |
ENSG00000222551 |
. |
50920 |
0.17 |
| chr3_66514964_66515132 | 0.26 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
36308 |
0.22 |
| chr5_79251776_79251927 | 0.26 |
RP11-168A11.1 |
|
19449 |
0.2 |
| chr6_49794571_49794884 | 0.26 |
RP3-417L20.4 |
|
9584 |
0.19 |
| chr1_39175768_39175919 | 0.26 |
RRAGC |
Ras-related GTP binding C |
149652 |
0.04 |
| chr19_33569394_33569545 | 0.26 |
GPATCH1 |
G patch domain containing 1 |
2317 |
0.26 |
| chr13_33778604_33778877 | 0.26 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
1403 |
0.49 |
| chr13_94069136_94069422 | 0.25 |
ENSG00000252365 |
. |
48066 |
0.2 |
| chr8_130734980_130735131 | 0.25 |
GSDMC |
gasdermin C |
64079 |
0.12 |
| chr13_96295623_96295774 | 0.25 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
181 |
0.96 |
| chr2_145765254_145765507 | 0.25 |
ENSG00000253036 |
. |
327258 |
0.01 |
| chr4_156363784_156363935 | 0.25 |
MAP9 |
microtubule-associated protein 9 |
65737 |
0.13 |
| chr17_73322605_73322792 | 0.24 |
GRB2 |
growth factor receptor-bound protein 2 |
4864 |
0.12 |
| chr18_25444147_25444298 | 0.24 |
AC015933.2 |
|
90261 |
0.1 |
| chr1_169014404_169014555 | 0.24 |
ENSG00000252987 |
. |
22023 |
0.19 |
| chr10_33527455_33527606 | 0.24 |
NRP1 |
neuropilin 1 |
25041 |
0.19 |
| chr4_123775694_123776122 | 0.24 |
ENSG00000253069 |
. |
27739 |
0.14 |
| chr7_115816824_115816975 | 0.24 |
TFEC |
transcription factor EC |
16949 |
0.21 |
| chr6_133006336_133006628 | 0.23 |
VNN1 |
vanin 1 |
28706 |
0.12 |
| chr1_229642144_229642295 | 0.23 |
NUP133 |
nucleoporin 133kDa |
1854 |
0.24 |
| chr3_177978802_177979100 | 0.23 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
11769 |
0.3 |
| chr15_60898079_60898454 | 0.22 |
RORA |
RAR-related orphan receptor A |
12941 |
0.23 |
| chr13_110064884_110065035 | 0.22 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
211128 |
0.02 |
| chr13_30419563_30419714 | 0.22 |
UBL3 |
ubiquitin-like 3 |
5183 |
0.32 |
| chr8_23397961_23398112 | 0.22 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
11463 |
0.16 |
| chrX_133804801_133804952 | 0.22 |
PLAC1 |
placenta-specific 1 |
12363 |
0.2 |
| chr10_122612352_122612503 | 0.22 |
WDR11 |
WD repeat domain 11 |
1428 |
0.37 |
| chr14_51910059_51910325 | 0.22 |
FRMD6 |
FERM domain containing 6 |
45663 |
0.16 |
| chr5_136751048_136751210 | 0.21 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
83118 |
0.1 |
| chr9_35688355_35688710 | 0.21 |
TPM2 |
tropomyosin 2 (beta) |
1368 |
0.2 |
| chr12_33050043_33050552 | 0.20 |
PKP2 |
plakophilin 2 |
523 |
0.87 |
| chr12_47898750_47898901 | 0.20 |
ENSG00000264906 |
. |
140773 |
0.04 |
| chr4_99214442_99214593 | 0.20 |
RP11-323J4.1 |
|
31736 |
0.19 |
| chr6_52924116_52924267 | 0.20 |
ICK |
intestinal cell (MAK-like) kinase |
2377 |
0.21 |
| chr8_18576139_18576290 | 0.20 |
RP11-161I2.1 |
|
2201 |
0.39 |
| chr10_99082888_99083039 | 0.20 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
3941 |
0.14 |
| chr18_20558047_20558453 | 0.20 |
RBBP8 |
retinoblastoma binding protein 8 |
9445 |
0.15 |
| chr2_12997455_12997766 | 0.19 |
ENSG00000264370 |
. |
120117 |
0.06 |
| chr1_68152763_68153190 | 0.19 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
2092 |
0.36 |
| chr3_156398302_156398550 | 0.19 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
529 |
0.81 |
| chr6_132524074_132524225 | 0.19 |
ENSG00000265669 |
. |
87746 |
0.09 |
| chr8_131013169_131013320 | 0.19 |
ENSG00000253043 |
. |
3603 |
0.21 |
| chr12_8088033_8088689 | 0.19 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
421 |
0.8 |
| chr8_22029446_22029597 | 0.19 |
BMP1 |
bone morphogenetic protein 1 |
6721 |
0.11 |
| chr20_10501395_10501546 | 0.19 |
SLX4IP |
SLX4 interacting protein |
85519 |
0.09 |
| chr12_82826657_82826808 | 0.19 |
METTL25 |
methyltransferase like 25 |
33594 |
0.21 |
| chr6_17993424_17993603 | 0.19 |
KIF13A |
kinesin family member 13A |
5659 |
0.3 |
| chr12_107617852_107618003 | 0.19 |
BTBD11 |
BTB (POZ) domain containing 11 |
94263 |
0.08 |
| chr12_104196646_104196797 | 0.18 |
RP11-650K20.2 |
|
16667 |
0.13 |
| chr20_19861450_19861601 | 0.18 |
RIN2 |
Ras and Rab interactor 2 |
5640 |
0.29 |
| chr2_75873758_75873909 | 0.18 |
MRPL19 |
mitochondrial ribosomal protein L19 |
76 |
0.95 |
| chr11_43902927_43903223 | 0.18 |
ALKBH3 |
alkB, alkylation repair homolog 3 (E. coli) |
670 |
0.47 |
| chr21_16431543_16432004 | 0.18 |
NRIP1 |
nuclear receptor interacting protein 1 |
5353 |
0.3 |
| chr17_71817273_71817424 | 0.18 |
SDK2 |
sidekick cell adhesion molecule 2 |
177120 |
0.03 |
| chr4_99576885_99577036 | 0.18 |
TSPAN5 |
tetraspanin 5 |
1826 |
0.38 |
| chr5_162930555_162931177 | 0.18 |
MAT2B |
methionine adenosyltransferase II, beta |
746 |
0.62 |
| chr2_123392168_123392319 | 0.18 |
TSN |
translin |
878496 |
0.0 |
| chr9_1173997_1174148 | 0.17 |
DMRT2 |
doublesex and mab-3 related transcription factor 2 |
122458 |
0.06 |
| chr21_17907841_17908039 | 0.17 |
ENSG00000207638 |
. |
3469 |
0.26 |
| chr4_89394952_89395117 | 0.17 |
HERC5 |
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
6027 |
0.17 |
| chr17_44805262_44805413 | 0.17 |
NSF |
N-ethylmaleimide-sensitive factor |
1415 |
0.5 |
| chrX_153023566_153023717 | 0.17 |
PLXNB3 |
plexin B3 |
6010 |
0.1 |
| chr8_119344904_119345055 | 0.17 |
AC023590.1 |
Uncharacterized protein |
50498 |
0.19 |
| chr1_201323781_201323932 | 0.17 |
TNNT2 |
troponin T type 2 (cardiac) |
18526 |
0.16 |
| chr15_33111539_33111777 | 0.17 |
FMN1 |
formin 1 |
68797 |
0.1 |
| chr1_36614255_36614499 | 0.17 |
TRAPPC3 |
trafficking protein particle complex 3 |
675 |
0.62 |
| chr6_139561908_139562059 | 0.17 |
RP1-225E12.2 |
|
1541 |
0.41 |
| chr2_207024982_207025205 | 0.17 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
762 |
0.32 |
| chr1_86969615_86969766 | 0.17 |
CLCA1 |
chloride channel accessory 1 |
35164 |
0.16 |
| chr3_183530706_183530857 | 0.17 |
YEATS2-AS1 |
YEATS2 antisense RNA 1 |
2210 |
0.22 |
| chr12_46659525_46659676 | 0.17 |
SLC38A1 |
solute carrier family 38, member 1 |
1884 |
0.48 |
| chr11_133450172_133450323 | 0.17 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
47833 |
0.17 |
| chr1_246646221_246646372 | 0.16 |
SMYD3 |
SET and MYND domain containing 3 |
771 |
0.7 |
| chr5_43310867_43311162 | 0.16 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
2312 |
0.32 |
| chr5_137674413_137674698 | 0.16 |
CDC25C |
cell division cycle 25C |
511 |
0.47 |
| chr10_134388453_134389007 | 0.16 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
32700 |
0.18 |
| chr5_173399860_173400035 | 0.16 |
C5orf47 |
chromosome 5 open reading frame 47 |
16215 |
0.22 |
| chr22_41253195_41254018 | 0.16 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
504 |
0.47 |
| chr10_13828862_13829013 | 0.16 |
RP11-353M9.1 |
|
57554 |
0.1 |
| chr2_201622052_201622203 | 0.16 |
ENSG00000201737 |
. |
18180 |
0.14 |
| chr6_21587871_21588206 | 0.16 |
SOX4 |
SRY (sex determining region Y)-box 4 |
5934 |
0.35 |
| chr3_106308061_106308212 | 0.16 |
ENSG00000200610 |
. |
73392 |
0.12 |
| chr22_19074199_19074350 | 0.16 |
AC004471.9 |
|
34768 |
0.09 |
| chr5_40490586_40490737 | 0.16 |
ENSG00000199552 |
. |
164404 |
0.03 |
| chr14_37131505_37132074 | 0.16 |
PAX9 |
paired box 9 |
10 |
0.98 |
| chr15_49913739_49914013 | 0.16 |
DTWD1 |
DTW domain containing 1 |
580 |
0.53 |
| chr4_11652300_11652527 | 0.16 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
221024 |
0.02 |
| chr12_109090441_109090824 | 0.16 |
CORO1C |
coronin, actin binding protein, 1C |
454 |
0.79 |
| chr3_95257506_95257657 | 0.16 |
ENSG00000221477 |
. |
628165 |
0.0 |
| chr1_192776419_192777001 | 0.15 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
1461 |
0.52 |
| chr9_127030053_127030414 | 0.15 |
RP11-121A14.3 |
|
5078 |
0.19 |
| chr4_105315936_105316087 | 0.15 |
ENSG00000272082 |
. |
96022 |
0.08 |
| chr3_37978259_37978410 | 0.15 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
27736 |
0.14 |
| chr6_162040062_162040271 | 0.15 |
ENSG00000221021 |
. |
4068 |
0.38 |
| chr1_94750293_94750444 | 0.15 |
ARHGAP29 |
Rho GTPase activating protein 29 |
47179 |
0.16 |
| chr2_75834857_75835085 | 0.15 |
ENSG00000221638 |
. |
20108 |
0.12 |
| chr10_47242799_47243052 | 0.15 |
BMS1P2 |
BMS1 pseudogene 2 |
1018 |
0.47 |
| chr1_89060919_89061070 | 0.15 |
RP11-76N22.2 |
|
24451 |
0.24 |
| chr20_44065109_44065260 | 0.15 |
AL031663.2 |
CDNA FLJ26875 fis, clone PRS08969; Uncharacterized protein |
8887 |
0.11 |
| chr7_7681092_7681243 | 0.15 |
RPA3 |
replication protein A3, 14kDa |
564 |
0.83 |
| chrX_105854737_105855044 | 0.15 |
CXorf57 |
chromosome X open reading frame 57 |
270 |
0.94 |
| chr11_66795405_66795556 | 0.15 |
SYT12 |
synaptotagmin XII |
449 |
0.77 |
| chrX_99904040_99904245 | 0.15 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
4927 |
0.19 |
| chr1_151513093_151513944 | 0.15 |
RP11-74C1.4 |
|
526 |
0.48 |
| chr2_85199284_85199564 | 0.15 |
KCMF1 |
potassium channel modulatory factor 1 |
100 |
0.98 |
| chr1_195606757_195606908 | 0.15 |
ENSG00000265108 |
. |
511021 |
0.0 |
| chrX_77040024_77040561 | 0.15 |
ATRX |
alpha thalassemia/mental retardation syndrome X-linked |
1410 |
0.49 |
| chr19_11096722_11096873 | 0.15 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
1969 |
0.25 |
| chr4_177710355_177710506 | 0.14 |
VEGFC |
vascular endothelial growth factor C |
3451 |
0.38 |
| chrX_13106517_13106711 | 0.14 |
FAM9C |
family with sequence similarity 9, member C |
43813 |
0.18 |
| chr11_34367200_34367440 | 0.14 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
11482 |
0.26 |
| chr12_57077777_57078354 | 0.14 |
PTGES3 |
prostaglandin E synthase 3 (cytosolic) |
3881 |
0.14 |
| chr1_184823664_184823815 | 0.14 |
ENSG00000252222 |
. |
33116 |
0.18 |
| chr20_10741569_10741720 | 0.14 |
JAG1 |
jagged 1 |
86950 |
0.09 |
| chrX_45182977_45183128 | 0.14 |
RP11-342D14.1 |
|
3349 |
0.38 |
| chr1_89457123_89457381 | 0.14 |
CCBL2 |
cysteine conjugate-beta lyase 2 |
1035 |
0.37 |
| chr6_53670019_53670170 | 0.14 |
LRRC1 |
leucine rich repeat containing 1 |
10206 |
0.21 |
| chr2_100427985_100428136 | 0.14 |
AFF3 |
AF4/FMR2 family, member 3 |
233228 |
0.02 |
| chr7_15725909_15726109 | 0.14 |
MEOX2 |
mesenchyme homeobox 2 |
428 |
0.88 |
| chr9_132404432_132404583 | 0.14 |
ASB6 |
ankyrin repeat and SOCS box containing 6 |
63 |
0.95 |
| chr6_21191491_21191642 | 0.14 |
ENSG00000252046 |
. |
197767 |
0.03 |
| chr11_115530687_115530852 | 0.14 |
ENSG00000239153 |
. |
32866 |
0.25 |
| chr3_160787530_160787756 | 0.14 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
31312 |
0.2 |
| chr1_32168571_32169014 | 0.14 |
COL16A1 |
collagen, type XVI, alpha 1 |
976 |
0.48 |
| chr2_40662150_40662301 | 0.14 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
4781 |
0.35 |
| chr15_78251949_78252110 | 0.14 |
ADAMTS7P3 |
ADAMTS7 pseudogene 3 |
16355 |
0.11 |
| chr3_58316326_58316512 | 0.14 |
PXK |
PX domain containing serine/threonine kinase |
2188 |
0.31 |
| chr4_170190845_170191352 | 0.14 |
SH3RF1 |
SH3 domain containing ring finger 1 |
10 |
0.99 |
| chr7_92241576_92241727 | 0.13 |
FAM133B |
family with sequence similarity 133, member B |
21943 |
0.2 |
| chr6_132713364_132713525 | 0.13 |
MOXD1 |
monooxygenase, DBH-like 1 |
9170 |
0.27 |
| chrX_40048176_40048340 | 0.13 |
BCOR |
BCL6 corepressor |
11676 |
0.29 |
| chr9_35746072_35746267 | 0.13 |
RGP1 |
RGP1 retrograde golgi transport homolog (S. cerevisiae) |
3034 |
0.1 |
| chr19_52873836_52874044 | 0.13 |
ZNF880 |
zinc finger protein 880 |
736 |
0.53 |
| chr2_242227382_242227533 | 0.13 |
HDLBP |
high density lipoprotein binding protein |
14682 |
0.14 |
| chr2_197089963_197090119 | 0.13 |
ENSG00000239161 |
. |
9995 |
0.19 |
| chr13_41765022_41765570 | 0.13 |
KBTBD7 |
kelch repeat and BTB (POZ) domain containing 7 |
3406 |
0.21 |
| chr6_12293530_12293681 | 0.13 |
EDN1 |
endothelin 1 |
3009 |
0.39 |
| chr18_55889704_55889927 | 0.13 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
1012 |
0.61 |
| chr12_94852826_94853545 | 0.13 |
CCDC41 |
coiled-coil domain containing 41 |
537 |
0.82 |
| chrX_149534374_149534815 | 0.13 |
MAMLD1 |
mastermind-like domain containing 1 |
2908 |
0.38 |
| chr9_113205949_113206100 | 0.13 |
SVEP1 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
15986 |
0.24 |
| chr20_9389767_9389966 | 0.13 |
PLCB4 |
phospholipase C, beta 4 |
101419 |
0.07 |
| chr7_41738474_41738677 | 0.13 |
INHBA |
inhibin, beta A |
1632 |
0.38 |
| chr3_4793867_4794159 | 0.13 |
ENSG00000239126 |
. |
126373 |
0.05 |
| chr1_61330866_61331017 | 0.13 |
NFIA |
nuclear factor I/A |
10 |
0.99 |
| chr9_107669806_107669965 | 0.13 |
RP11-217B7.2 |
|
19949 |
0.21 |
| chr11_27492592_27492931 | 0.13 |
RP11-159H22.2 |
|
515 |
0.65 |
| chr12_48082357_48082508 | 0.13 |
RPAP3 |
RNA polymerase II associated protein 3 |
17337 |
0.17 |
| chr3_114865060_114865714 | 0.13 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
731 |
0.81 |
| chr11_131618597_131618988 | 0.13 |
NTM |
neurotrimin |
87904 |
0.1 |
| chr5_75842727_75842878 | 0.13 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
432 |
0.88 |
| chr19_16195500_16195773 | 0.13 |
TPM4 |
tropomyosin 4 |
3103 |
0.21 |
| chr6_118971873_118972263 | 0.13 |
CEP85L |
centrosomal protein 85kDa-like |
952 |
0.71 |
| chr6_36590990_36591141 | 0.13 |
ENSG00000263894 |
. |
776 |
0.56 |
| chr5_17805402_17805553 | 0.13 |
ENSG00000212278 |
. |
430755 |
0.01 |
| chr1_225968168_225968319 | 0.13 |
SRP9 |
signal recognition particle 9kDa |
2606 |
0.22 |
| chr18_9138515_9138757 | 0.13 |
ANKRD12 |
ankyrin repeat domain 12 |
1075 |
0.42 |
| chr8_76319339_76319966 | 0.13 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
619 |
0.85 |
| chr4_83280489_83280640 | 0.13 |
HNRNPD |
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
228 |
0.93 |
| chr4_157867457_157867608 | 0.13 |
PDGFC |
platelet derived growth factor C |
24523 |
0.2 |
| chr1_109583292_109583606 | 0.13 |
WDR47 |
WD repeat domain 47 |
720 |
0.6 |
| chr12_46762957_46763490 | 0.12 |
RP11-474P2.2 |
|
2023 |
0.36 |
| chr1_146640938_146641089 | 0.12 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
3033 |
0.2 |
| chr5_36664801_36664952 | 0.12 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
56445 |
0.14 |
| chr12_78683757_78683908 | 0.12 |
NAV3 |
neuron navigator 3 |
90976 |
0.1 |
| chr2_201270113_201270684 | 0.12 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
12598 |
0.18 |
| chr5_95170159_95170387 | 0.12 |
GLRX |
glutaredoxin (thioltransferase) |
11564 |
0.14 |
| chr4_166252557_166253372 | 0.12 |
MSMO1 |
methylsterol monooxygenase 1 |
3885 |
0.25 |
| chr4_169490767_169491028 | 0.12 |
PALLD |
palladin, cytoskeletal associated protein |
58178 |
0.12 |
| chr18_56215999_56216150 | 0.12 |
RP11-126O1.2 |
|
8228 |
0.17 |
| chrX_57622345_57622884 | 0.12 |
ZXDB |
zinc finger, X-linked, duplicated B |
4345 |
0.37 |
| chr4_138281360_138281511 | 0.12 |
ENSG00000222526 |
. |
132153 |
0.06 |
| chr17_46048416_46049087 | 0.12 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
234 |
0.85 |
| chr7_41426318_41426600 | 0.12 |
INHBA-AS1 |
INHBA antisense RNA 1 |
307055 |
0.01 |
| chrX_33225807_33225958 | 0.12 |
DMD |
dystrophin |
3547 |
0.4 |
| chr11_100761786_100762126 | 0.12 |
ARHGAP42 |
Rho GTPase activating protein 42 |
22275 |
0.19 |
| chr15_56204998_56205389 | 0.12 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
2659 |
0.34 |
| chr1_211056531_211056682 | 0.12 |
KCNH1-IT1 |
KCNH1 intronic transcript 1 (non-protein coding) |
250110 |
0.02 |
| chr15_63448807_63449350 | 0.12 |
RPS27L |
ribosomal protein S27-like |
105 |
0.97 |
| chr2_145153762_145153913 | 0.12 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
33756 |
0.21 |
| chr1_159819723_159819925 | 0.12 |
ENSG00000212161 |
. |
1938 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:1902808 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |