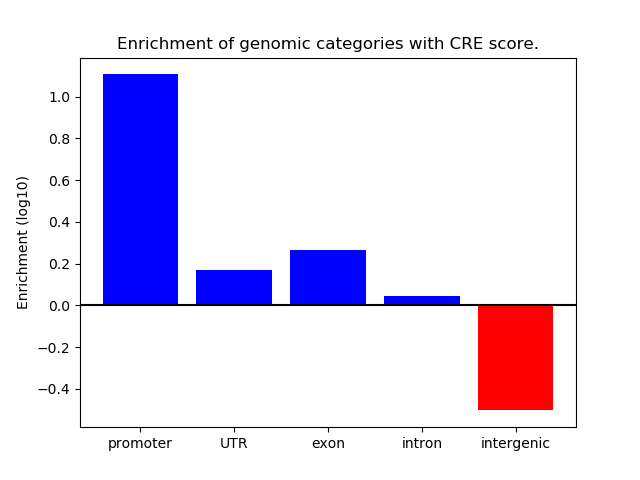
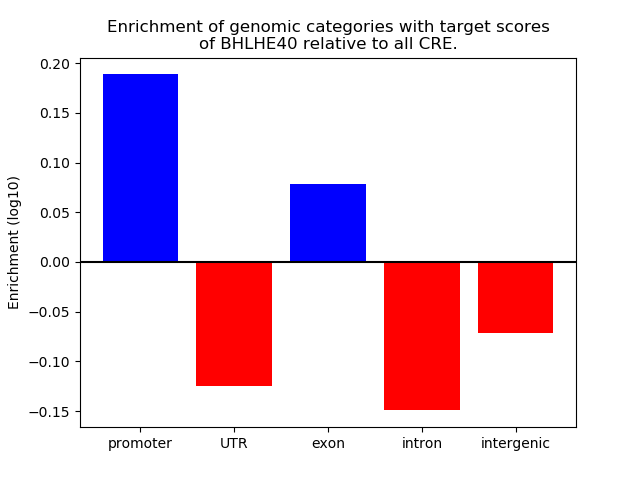
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
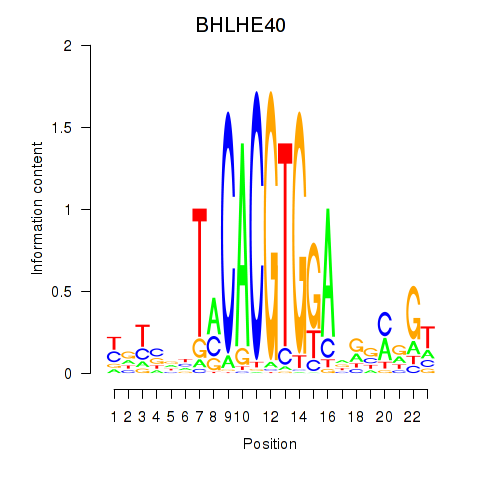
Results for BHLHE40
Z-value: 0.78
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_4992051_4992202 | BHLHE40 | 28675 | 0.163351 | -0.71 | 3.1e-02 | Click! |
| chr3_5019013_5019527 | BHLHE40 | 1531 | 0.376496 | -0.67 | 4.7e-02 | Click! |
| chr3_5019531_5019715 | BHLHE40 | 1178 | 0.450298 | -0.32 | 4.1e-01 | Click! |
| chr3_5020528_5020679 | BHLHE40 | 198 | 0.899685 | -0.26 | 5.0e-01 | Click! |
| chr3_5019761_5020512 | BHLHE40 | 665 | 0.639724 | -0.03 | 9.3e-01 | Click! |
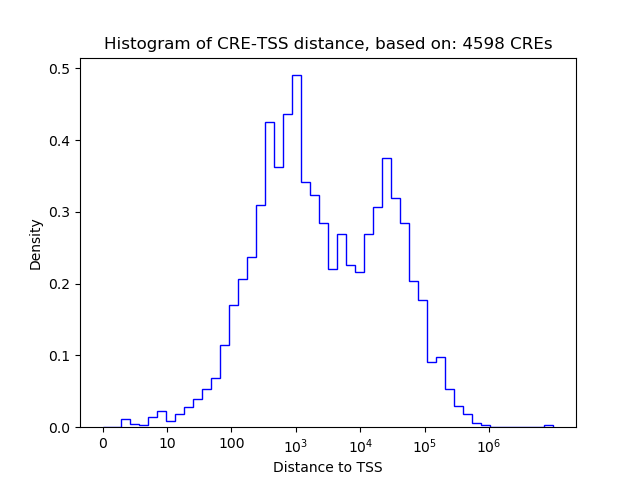
Activity of the BHLHE40 motif across conditions
Conditions sorted by the z-value of the BHLHE40 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
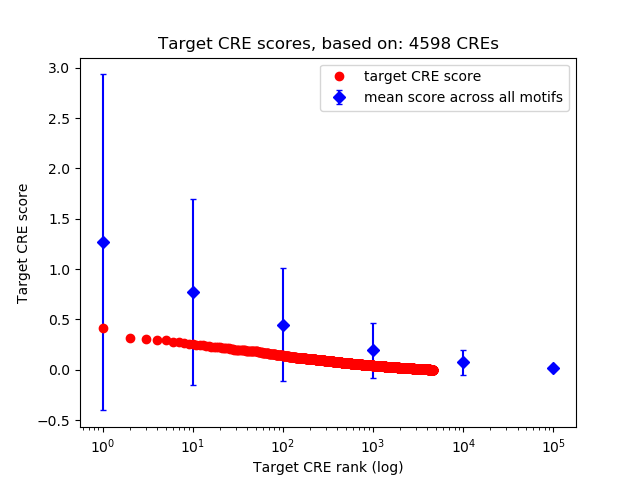
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_2077067_2078193 | 0.41 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
701 |
0.35 |
| chr10_128076215_128076453 | 0.31 |
ADAM12 |
ADAM metallopeptidase domain 12 |
690 |
0.79 |
| chr13_102105916_102106117 | 0.30 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
998 |
0.66 |
| chr1_68297532_68297957 | 0.30 |
GNG12-AS1 |
GNG12 antisense RNA 1 |
242 |
0.9 |
| chr22_25507059_25507657 | 0.30 |
CTA-221G9.11 |
|
1301 |
0.51 |
| chr10_63809082_63810748 | 0.27 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
945 |
0.69 |
| chr3_36422437_36422689 | 0.27 |
STAC |
SH3 and cysteine rich domain |
498 |
0.89 |
| chr4_114899856_114900283 | 0.27 |
ARSJ |
arylsulfatase family, member J |
83 |
0.98 |
| chr12_124872056_124872210 | 0.26 |
NCOR2 |
nuclear receptor corepressor 2 |
1237 |
0.6 |
| chr10_112113035_112113246 | 0.25 |
SMNDC1 |
survival motor neuron domain containing 1 |
48431 |
0.14 |
| chr2_12860250_12860601 | 0.25 |
TRIB2 |
tribbles pseudokinase 2 |
2061 |
0.41 |
| chr3_197326133_197326372 | 0.24 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
26058 |
0.17 |
| chr8_101859554_101859783 | 0.24 |
ENSG00000222795 |
. |
15580 |
0.19 |
| chr17_29887977_29888214 | 0.24 |
ENSG00000207614 |
. |
1080 |
0.35 |
| chr1_86047684_86048329 | 0.24 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
1562 |
0.37 |
| chr9_21989659_21989874 | 0.23 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
4644 |
0.16 |
| chr18_46473562_46473713 | 0.23 |
SMAD7 |
SMAD family member 7 |
1238 |
0.56 |
| chr21_46727414_46727565 | 0.22 |
ENSG00000215447 |
. |
19522 |
0.16 |
| chr15_60682035_60682438 | 0.22 |
ANXA2 |
annexin A2 |
1090 |
0.63 |
| chrX_17395110_17395408 | 0.22 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
1716 |
0.4 |
| chr8_145046455_145046834 | 0.22 |
PLEC |
plectin |
1053 |
0.34 |
| chr7_116166933_116167162 | 0.22 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
700 |
0.64 |
| chr11_69460305_69460629 | 0.22 |
CCND1 |
cyclin D1 |
4493 |
0.24 |
| chr11_118791642_118792157 | 0.21 |
BCL9L |
B-cell CLL/lymphoma 9-like |
2286 |
0.14 |
| chr21_47519356_47519507 | 0.21 |
COL6A2 |
collagen, type VI, alpha 2 |
720 |
0.56 |
| chr10_52176688_52176970 | 0.21 |
AC069547.2 |
Uncharacterized protein |
24063 |
0.19 |
| chr8_494269_494491 | 0.21 |
TDRP |
testis development related protein |
457 |
0.88 |
| chr4_20256302_20256578 | 0.20 |
SLIT2 |
slit homolog 2 (Drosophila) |
103 |
0.98 |
| chr13_100636352_100636503 | 0.20 |
ZIC2 |
Zic family member 2 |
2401 |
0.28 |
| chr3_52567337_52567575 | 0.20 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
91 |
0.94 |
| chr9_130737928_130738103 | 0.20 |
FAM102A |
family with sequence similarity 102, member A |
4777 |
0.13 |
| chr5_179778925_179779076 | 0.20 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
900 |
0.63 |
| chr11_114168914_114169158 | 0.20 |
NNMT |
nicotinamide N-methyltransferase |
163 |
0.96 |
| chr13_34184631_34184800 | 0.20 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
207471 |
0.02 |
| chr17_40932934_40933198 | 0.20 |
WNK4 |
WNK lysine deficient protein kinase 4 |
370 |
0.71 |
| chr7_94028286_94028494 | 0.20 |
COL1A2 |
collagen, type I, alpha 2 |
4517 |
0.32 |
| chr17_74697030_74697413 | 0.20 |
ENSG00000212418 |
. |
2588 |
0.13 |
| chr7_42275396_42275690 | 0.19 |
GLI3 |
GLI family zinc finger 3 |
1069 |
0.7 |
| chr5_73839584_73839978 | 0.19 |
HEXB |
hexosaminidase B (beta polypeptide) |
96067 |
0.08 |
| chr9_35491110_35491278 | 0.19 |
RUSC2 |
RUN and SH3 domain containing 2 |
1070 |
0.5 |
| chr16_86695305_86695456 | 0.19 |
FOXL1 |
forkhead box L1 |
83265 |
0.09 |
| chr5_98111445_98111873 | 0.19 |
RGMB |
repulsive guidance molecule family member b |
2320 |
0.3 |
| chr2_145594298_145594531 | 0.19 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
315793 |
0.01 |
| chr6_143259748_143259920 | 0.19 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
6504 |
0.28 |
| chrX_105970264_105970617 | 0.19 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
546 |
0.83 |
| chr16_11422925_11423155 | 0.19 |
RMI2 |
RecQ mediated genome instability 2 |
12404 |
0.08 |
| chr14_42074686_42074934 | 0.19 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
1963 |
0.41 |
| chr15_85525955_85526270 | 0.19 |
PDE8A |
phosphodiesterase 8A |
900 |
0.6 |
| chr2_72370559_72370716 | 0.18 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
424 |
0.92 |
| chr11_79150433_79150655 | 0.18 |
TENM4 |
teneurin transmembrane protein 4 |
1151 |
0.54 |
| chr1_32169073_32169273 | 0.18 |
COL16A1 |
collagen, type XVI, alpha 1 |
595 |
0.68 |
| chr2_216301575_216301809 | 0.18 |
AC012462.1 |
|
716 |
0.48 |
| chr15_68723417_68723663 | 0.18 |
ITGA11 |
integrin, alpha 11 |
952 |
0.69 |
| chr20_57426029_57426307 | 0.18 |
GNAS-AS1 |
GNAS antisense RNA 1 |
210 |
0.89 |
| chr12_49318526_49318771 | 0.18 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
102 |
0.91 |
| chr19_6531750_6531901 | 0.18 |
TNFSF9 |
tumor necrosis factor (ligand) superfamily, member 9 |
815 |
0.47 |
| chr8_37350120_37350468 | 0.18 |
RP11-150O12.6 |
|
24245 |
0.24 |
| chr9_98272289_98272560 | 0.18 |
PTCH1 |
patched 1 |
1481 |
0.34 |
| chr2_151337593_151337891 | 0.17 |
RND3 |
Rho family GTPase 3 |
4154 |
0.38 |
| chr5_121648324_121648736 | 0.17 |
SNCAIP |
synuclein, alpha interacting protein |
268 |
0.87 |
| chr10_44069814_44070416 | 0.17 |
ZNF239 |
zinc finger protein 239 |
49 |
0.98 |
| chr5_54281245_54281752 | 0.17 |
ESM1 |
endothelial cell-specific molecule 1 |
7 |
0.97 |
| chr9_2157555_2157766 | 0.17 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
20 |
0.98 |
| chr17_32906311_32906556 | 0.17 |
C17orf102 |
chromosome 17 open reading frame 102 |
45 |
0.97 |
| chr1_56441058_56441209 | 0.16 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
36180 |
0.24 |
| chr20_31062458_31062723 | 0.16 |
C20orf112 |
chromosome 20 open reading frame 112 |
8684 |
0.18 |
| chr11_133917712_133918072 | 0.16 |
JAM3 |
junctional adhesion molecule 3 |
20928 |
0.21 |
| chr12_131322718_131323133 | 0.16 |
STX2 |
syntaxin 2 |
814 |
0.6 |
| chr5_139724897_139725179 | 0.16 |
HBEGF |
heparin-binding EGF-like growth factor |
1150 |
0.36 |
| chr14_99740123_99740356 | 0.16 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
2378 |
0.32 |
| chr14_52121858_52122101 | 0.16 |
FRMD6 |
FERM domain containing 6 |
3281 |
0.25 |
| chr12_125395384_125395562 | 0.16 |
UBC |
ubiquitin C |
3147 |
0.2 |
| chrX_17609979_17610432 | 0.16 |
NHS-AS1 |
NHS antisense RNA 1 |
34697 |
0.17 |
| chr5_136645949_136646125 | 0.16 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
3190 |
0.39 |
| chr4_99575076_99575227 | 0.16 |
TSPAN5 |
tetraspanin 5 |
3635 |
0.27 |
| chr5_140857632_140858335 | 0.16 |
ENSG00000242020 |
. |
900 |
0.34 |
| chr6_132271257_132271718 | 0.16 |
RP11-69I8.3 |
|
599 |
0.62 |
| chr7_142507018_142507186 | 0.15 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
25971 |
0.15 |
| chr1_85665779_85666437 | 0.15 |
SYDE2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
621 |
0.72 |
| chr10_93058252_93058702 | 0.15 |
PCGF5 |
polycomb group ring finger 5 |
77932 |
0.11 |
| chr3_116163640_116163792 | 0.15 |
LSAMP |
limbic system-associated membrane protein |
114 |
0.98 |
| chr16_73266705_73266923 | 0.15 |
C16orf47 |
chromosome 16 open reading frame 47 |
88468 |
0.1 |
| chr17_1081882_1082496 | 0.15 |
ABR |
active BCR-related |
889 |
0.58 |
| chr3_47051058_47051537 | 0.15 |
NBEAL2 |
neurobeachin-like 2 |
4526 |
0.22 |
| chr11_469322_469620 | 0.15 |
RP13-317D12.3 |
|
5572 |
0.1 |
| chr1_42383823_42384248 | 0.15 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
134 |
0.98 |
| chr7_27940568_27941235 | 0.15 |
ENSG00000265382 |
. |
20795 |
0.23 |
| chr7_73133202_73133898 | 0.15 |
STX1A |
syntaxin 1A (brain) |
411 |
0.72 |
| chr10_13932932_13933715 | 0.15 |
FRMD4A |
FERM domain containing 4A |
32411 |
0.2 |
| chr22_46462679_46462866 | 0.15 |
RP6-109B7.4 |
|
2999 |
0.14 |
| chr9_130616400_130616906 | 0.15 |
ENG |
endoglin |
262 |
0.8 |
| chr2_241376186_241376470 | 0.15 |
GPC1 |
glypican 1 |
1240 |
0.43 |
| chr1_40960645_40960796 | 0.15 |
RP11-656D10.3 |
|
13613 |
0.1 |
| chr21_35319646_35320126 | 0.15 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
1144 |
0.46 |
| chr1_162532826_162532977 | 0.14 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
1578 |
0.3 |
| chr16_10934760_10934911 | 0.14 |
TVP23A |
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
22193 |
0.15 |
| chr4_147560181_147560405 | 0.14 |
ENSG00000264323 |
. |
120 |
0.68 |
| chr5_158488903_158489314 | 0.14 |
EBF1 |
early B-cell factor 1 |
37593 |
0.17 |
| chr12_8814552_8814785 | 0.14 |
MFAP5 |
microfibrillar associated protein 5 |
1 |
0.97 |
| chr8_24799151_24799367 | 0.14 |
CTD-2168K21.2 |
|
14876 |
0.17 |
| chr5_159214342_159214493 | 0.14 |
ADRA1B |
adrenoceptor alpha 1B |
129373 |
0.05 |
| chr8_72459619_72459990 | 0.14 |
RP11-1102P16.1 |
Uncharacterized protein |
88 |
0.98 |
| chr4_7907648_7907905 | 0.14 |
AC097381.1 |
|
32952 |
0.16 |
| chr5_37839189_37839372 | 0.14 |
GDNF |
glial cell derived neurotrophic factor |
502 |
0.85 |
| chr5_146889201_146889394 | 0.14 |
DPYSL3 |
dihydropyrimidinase-like 3 |
322 |
0.94 |
| chr20_62372328_62372826 | 0.14 |
SLC2A4RG |
SLC2A4 regulator |
1363 |
0.22 |
| chr6_169652381_169652976 | 0.14 |
THBS2 |
thrombospondin 2 |
432 |
0.89 |
| chr1_46597349_46597672 | 0.14 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
778 |
0.56 |
| chr13_58203621_58204059 | 0.14 |
PCDH17 |
protocadherin 17 |
2815 |
0.43 |
| chr19_51433227_51433378 | 0.14 |
KLK4 |
kallikrein-related peptidase 4 |
19308 |
0.07 |
| chr17_47210248_47210565 | 0.14 |
B4GALNT2 |
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
77 |
0.97 |
| chr3_32021239_32021390 | 0.14 |
OSBPL10 |
oxysterol binding protein-like 10 |
1478 |
0.36 |
| chrX_135230110_135230418 | 0.13 |
FHL1 |
four and a half LIM domains 1 |
6 |
0.98 |
| chr14_29242746_29242955 | 0.13 |
C14orf23 |
chromosome 14 open reading frame 23 |
849 |
0.55 |
| chr6_28456937_28457274 | 0.13 |
GPX6 |
glutathione peroxidase 6 (olfactory) |
26459 |
0.14 |
| chr14_99655246_99655958 | 0.13 |
AL162151.4 |
|
30849 |
0.2 |
| chr7_150716014_150716204 | 0.13 |
ATG9B |
autophagy related 9B |
5477 |
0.1 |
| chr18_55863472_55863729 | 0.13 |
RP11-718I15.1 |
|
672 |
0.47 |
| chr11_120080847_120081389 | 0.13 |
OAF |
OAF homolog (Drosophila) |
357 |
0.86 |
| chr12_39836327_39837104 | 0.13 |
KIF21A |
kinesin family member 21A |
57 |
0.7 |
| chr20_52839047_52839198 | 0.13 |
PFDN4 |
prefoldin subunit 4 |
14736 |
0.25 |
| chr17_52978401_52978600 | 0.13 |
TOM1L1 |
target of myb1 (chicken)-like 1 |
5 |
0.99 |
| chr4_142693171_142693378 | 0.13 |
IL15 |
interleukin 15 |
50972 |
0.19 |
| chr12_52257745_52258098 | 0.13 |
ANKRD33 |
ankyrin repeat domain 33 |
23823 |
0.16 |
| chr11_66090538_66090689 | 0.13 |
CD248 |
CD248 molecule, endosialin |
6098 |
0.07 |
| chr8_96036821_96037145 | 0.13 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
231 |
0.92 |
| chr16_1233245_1233443 | 0.13 |
RP11-616M22.2 |
|
23168 |
0.08 |
| chr12_59311566_59311750 | 0.13 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1669 |
0.41 |
| chr6_133890397_133891036 | 0.12 |
RP3-323P13.2 |
|
57850 |
0.15 |
| chr12_3278190_3278426 | 0.12 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
19345 |
0.21 |
| chr8_1771644_1772131 | 0.12 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
255 |
0.93 |
| chr1_23751694_23751845 | 0.12 |
TCEA3 |
transcription elongation factor A (SII), 3 |
536 |
0.73 |
| chrX_66764418_66765028 | 0.12 |
AR |
androgen receptor |
61 |
0.99 |
| chr4_143300832_143301092 | 0.12 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
51450 |
0.19 |
| chr11_10679210_10679389 | 0.12 |
MRVI1 |
murine retrovirus integration site 1 homolog |
5376 |
0.21 |
| chr17_2077835_2078037 | 0.12 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
1464 |
0.31 |
| chr15_73979338_73979524 | 0.12 |
CD276 |
CD276 molecule |
2084 |
0.37 |
| chr22_35747250_35747434 | 0.12 |
ENSG00000266320 |
. |
15709 |
0.16 |
| chr9_22006120_22006693 | 0.12 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
2546 |
0.22 |
| chrX_54509628_54509782 | 0.12 |
FGD1 |
FYVE, RhoGEF and PH domain containing 1 |
12894 |
0.21 |
| chr13_74710745_74710896 | 0.12 |
KLF12 |
Kruppel-like factor 12 |
2426 |
0.45 |
| chr9_95970167_95970323 | 0.12 |
WNK2 |
WNK lysine deficient protein kinase 2 |
23020 |
0.19 |
| chr16_73097793_73098198 | 0.12 |
ZFHX3 |
zinc finger homeobox 3 |
4398 |
0.27 |
| chr6_28410806_28411495 | 0.12 |
ZSCAN23 |
zinc finger and SCAN domain containing 23 |
94 |
0.96 |
| chr9_98269032_98269993 | 0.12 |
PTCH1 |
patched 1 |
31 |
0.95 |
| chr2_121692732_121692961 | 0.12 |
ENSG00000221321 |
. |
99803 |
0.08 |
| chr4_20252886_20253037 | 0.12 |
SLIT2 |
slit homolog 2 (Drosophila) |
1922 |
0.5 |
| chr6_1554751_1555484 | 0.12 |
ENSG00000243439 |
. |
47560 |
0.16 |
| chr2_216289836_216290038 | 0.12 |
FN1 |
fibronectin 1 |
10853 |
0.22 |
| chr5_172160432_172160661 | 0.12 |
RP11-779O18.1 |
|
24683 |
0.12 |
| chr3_126853954_126854422 | 0.12 |
C3orf56 |
chromosome 3 open reading frame 56 |
57786 |
0.13 |
| chr1_151966472_151966834 | 0.12 |
S100A10 |
S100 calcium binding protein A10 |
213 |
0.65 |
| chr3_115507533_115507684 | 0.12 |
ENSG00000243359 |
. |
49107 |
0.19 |
| chr17_77788750_77788949 | 0.12 |
ENSG00000238331 |
. |
4371 |
0.16 |
| chr1_27357912_27358708 | 0.12 |
FAM46B |
family with sequence similarity 46, member B |
18983 |
0.16 |
| chr5_155109546_155109697 | 0.12 |
ENSG00000200275 |
. |
162826 |
0.04 |
| chr1_70509689_70509840 | 0.12 |
RP11-181B18.1 |
|
12859 |
0.27 |
| chr1_53664021_53664172 | 0.12 |
CPT2 |
carnitine palmitoyltransferase 2 |
1995 |
0.2 |
| chr16_29912249_29912746 | 0.12 |
ASPHD1 |
aspartate beta-hydroxylase domain containing 1 |
456 |
0.59 |
| chr19_10542095_10542754 | 0.12 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
687 |
0.54 |
| chr2_239770660_239770811 | 0.12 |
TWIST2 |
twist family bHLH transcription factor 2 |
14062 |
0.25 |
| chr16_15529020_15529579 | 0.12 |
C16orf45 |
chromosome 16 open reading frame 45 |
967 |
0.56 |
| chr22_46480425_46480827 | 0.12 |
FLJ27365 |
hsa-mir-4763 |
1257 |
0.28 |
| chr5_114795809_114796168 | 0.12 |
FEM1C |
fem-1 homolog c (C. elegans) |
84603 |
0.08 |
| chr19_48893644_48893831 | 0.12 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
375 |
0.76 |
| chr1_6557689_6557891 | 0.12 |
PLEKHG5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
306 |
0.84 |
| chr15_96877328_96877479 | 0.12 |
ENSG00000222651 |
. |
913 |
0.39 |
| chr3_184036946_184037180 | 0.12 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
76 |
0.94 |
| chr9_101557838_101558540 | 0.11 |
ANKS6 |
ankyrin repeat and sterile alpha motif domain containing 6 |
605 |
0.76 |
| chr19_44285188_44285480 | 0.11 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
75 |
0.96 |
| chr8_132053641_132053792 | 0.11 |
ADCY8 |
adenylate cyclase 8 (brain) |
956 |
0.71 |
| chr6_169889311_169889462 | 0.11 |
WDR27 |
WD repeat domain 27 |
171402 |
0.03 |
| chr15_29396229_29396380 | 0.11 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
59777 |
0.14 |
| chrX_30906152_30906450 | 0.11 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
20982 |
0.19 |
| chr12_49660713_49661148 | 0.11 |
TUBA1C |
tubulin, alpha 1c |
2066 |
0.18 |
| chr20_55207246_55207474 | 0.11 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
1422 |
0.37 |
| chr19_13166352_13166593 | 0.11 |
AC007787.2 |
|
15716 |
0.1 |
| chr11_130641825_130641976 | 0.11 |
C11orf44 |
chromosome 11 open reading frame 44 |
99049 |
0.08 |
| chr14_23013027_23013178 | 0.11 |
AE000662.92 |
Uncharacterized protein |
12432 |
0.1 |
| chr5_980906_981122 | 0.11 |
RP11-661C8.3 |
Uncharacterized protein |
1534 |
0.31 |
| chr14_90097002_90097153 | 0.11 |
RP11-944C7.1 |
Protein LOC100506792 |
833 |
0.53 |
| chr12_6876888_6877289 | 0.11 |
MLF2 |
myeloid leukemia factor 2 |
447 |
0.58 |
| chr2_45429_45675 | 0.11 |
FAM110C |
family with sequence similarity 110, member C |
833 |
0.76 |
| chr15_32267489_32267710 | 0.11 |
ENSG00000206849 |
. |
47093 |
0.15 |
| chr6_158034126_158034277 | 0.11 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
20160 |
0.25 |
| chr19_35757937_35758321 | 0.11 |
USF2 |
upstream transcription factor 2, c-fos interacting |
1752 |
0.18 |
| chr3_190580567_190581193 | 0.11 |
GMNC |
geminin coiled-coil domain containing |
476 |
0.9 |
| chr1_36180862_36181228 | 0.11 |
C1orf216 |
chromosome 1 open reading frame 216 |
4028 |
0.18 |
| chr7_97841044_97841241 | 0.11 |
BHLHA15 |
basic helix-loop-helix family, member a15 |
403 |
0.86 |
| chr3_29327052_29327571 | 0.11 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
4204 |
0.26 |
| chr14_96558753_96558904 | 0.11 |
C14orf132 |
chromosome 14 open reading frame 132 |
2232 |
0.34 |
| chr19_23185776_23185988 | 0.11 |
ZNF728 |
zinc finger protein 728 |
96 |
0.98 |
| chr12_50067178_50067329 | 0.11 |
FMNL3 |
formin-like 3 |
33750 |
0.11 |
| chr9_137437911_137438062 | 0.11 |
COL5A1 |
collagen, type V, alpha 1 |
95634 |
0.07 |
| chr18_77376610_77376761 | 0.11 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
63116 |
0.12 |
| chr9_130328652_130328977 | 0.11 |
FAM129B |
family with sequence similarity 129, member B |
2553 |
0.26 |
| chr9_44867829_44868127 | 0.11 |
RP11-160N1.10 |
|
407 |
0.89 |
| chr10_64323835_64324021 | 0.11 |
ZNF365 |
zinc finger protein 365 |
43721 |
0.19 |
| chrX_139847263_139847516 | 0.11 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
19334 |
0.22 |
| chrX_64254778_64255285 | 0.11 |
ZC4H2 |
zinc finger, C4H2 domain containing |
438 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.2 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.1 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.2 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0072071 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0051280 | negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.2 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0060412 | ventricular septum morphogenesis(GO:0060412) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0032356 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |