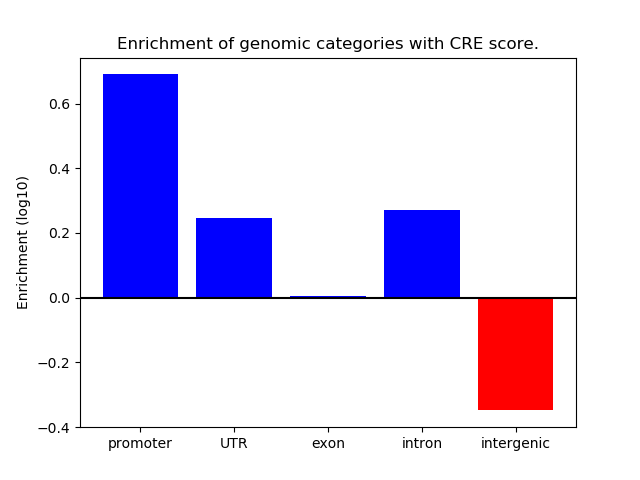
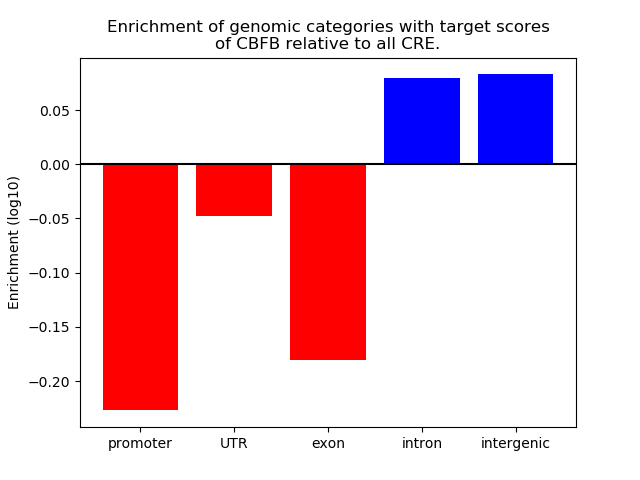
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
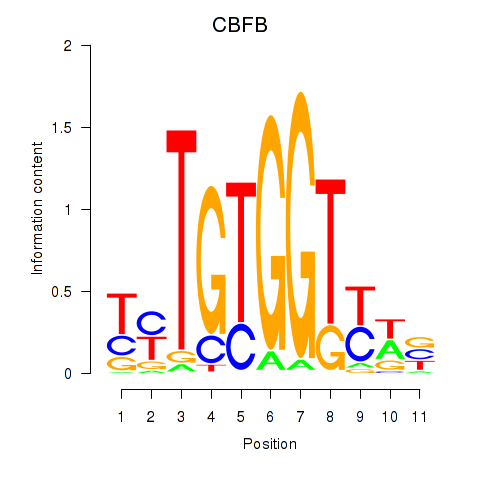
Results for CBFB
Z-value: 0.23
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
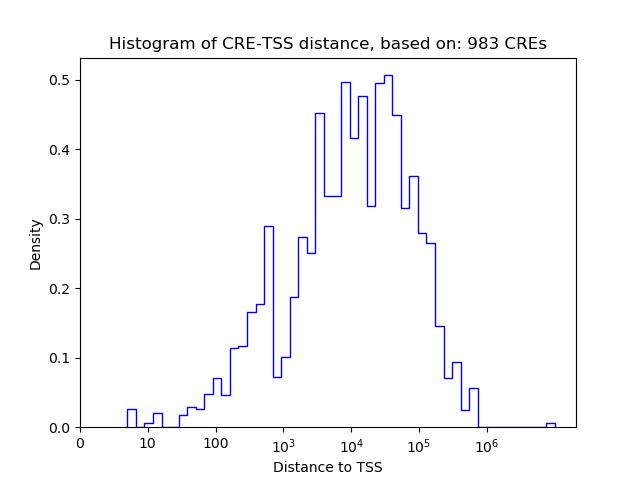
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_67063172_67063793 | CBFB | 118 | 0.942106 | -0.62 | 7.4e-02 | Click! |
| chr16_67096966_67097117 | CBFB | 33186 | 0.087619 | 0.52 | 1.5e-01 | Click! |
| chr16_67089936_67090087 | CBFB | 26156 | 0.098751 | 0.46 | 2.1e-01 | Click! |
| chr16_67066013_67066164 | CBFB | 2233 | 0.200612 | -0.43 | 2.5e-01 | Click! |
| chr16_67062127_67062421 | CBFB | 745 | 0.533375 | -0.36 | 3.5e-01 | Click! |
Activity of the CBFB motif across conditions
Conditions sorted by the z-value of the CBFB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
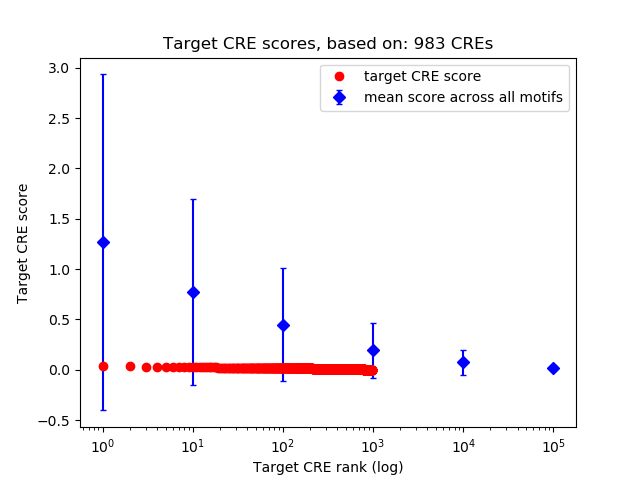
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_44347613_44347791 | 0.04 |
ENSG00000252599 |
. |
34332 |
0.15 |
| chr18_74843262_74844680 | 0.04 |
MBP |
myelin basic protein |
331 |
0.94 |
| chr16_55542879_55544136 | 0.03 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
597 |
0.77 |
| chr16_89392054_89392556 | 0.03 |
AC137932.6 |
|
2673 |
0.19 |
| chr1_9953446_9953936 | 0.03 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
396 |
0.8 |
| chr7_19144831_19144982 | 0.03 |
AC003986.6 |
|
7191 |
0.17 |
| chr10_88159807_88161210 | 0.03 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
34273 |
0.17 |
| chr2_55644066_55644508 | 0.03 |
CCDC88A |
coiled-coil domain containing 88A |
2125 |
0.27 |
| chr9_7976512_7976663 | 0.02 |
TMEM261 |
transmembrane protein 261 |
176520 |
0.04 |
| chr13_28975822_28976139 | 0.02 |
FLT1 |
fms-related tyrosine kinase 1 |
55554 |
0.16 |
| chr9_18976648_18977105 | 0.02 |
FAM154A |
family with sequence similarity 154, member A |
56310 |
0.1 |
| chr1_209823926_209824302 | 0.02 |
LAMB3 |
laminin, beta 3 |
565 |
0.7 |
| chr4_124981640_124982053 | 0.02 |
ANKRD50 |
ankyrin repeat domain 50 |
652041 |
0.0 |
| chr15_90380123_90380631 | 0.02 |
ENSG00000264966 |
. |
13576 |
0.11 |
| chr8_106102434_106102585 | 0.02 |
RP11-574O7.1 |
|
98325 |
0.09 |
| chr12_112818867_112820166 | 0.02 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
380 |
0.87 |
| chr3_49394601_49395955 | 0.02 |
GPX1 |
glutathione peroxidase 1 |
438 |
0.72 |
| chr13_68090205_68090367 | 0.02 |
PCDH9 |
protocadherin 9 |
285818 |
0.01 |
| chr11_122029721_122030081 | 0.02 |
ENSG00000207994 |
. |
6885 |
0.18 |
| chr17_20491916_20492779 | 0.02 |
CDRT15L2 |
CMT1A duplicated region transcript 15-like 2 |
9310 |
0.21 |
| chr1_8433877_8434267 | 0.02 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
9174 |
0.19 |
| chr5_166887847_166887998 | 0.02 |
CTB-105L4.2 |
|
151047 |
0.04 |
| chr6_154490671_154490822 | 0.02 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
77244 |
0.11 |
| chr10_98272514_98273588 | 0.02 |
TLL2 |
tolloid-like 2 |
617 |
0.79 |
| chr6_165722551_165723097 | 0.02 |
C6orf118 |
chromosome 6 open reading frame 118 |
272 |
0.95 |
| chr9_94182057_94182286 | 0.02 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
3973 |
0.34 |
| chr17_57719590_57719741 | 0.02 |
CLTC |
clathrin, heavy chain (Hc) |
22372 |
0.17 |
| chr12_41583641_41584059 | 0.02 |
PDZRN4 |
PDZ domain containing ring finger 4 |
1600 |
0.56 |
| chr10_130673966_130674117 | 0.02 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
591407 |
0.0 |
| chr4_153527366_153527517 | 0.02 |
RP11-768B22.2 |
|
60079 |
0.11 |
| chr3_31402829_31403285 | 0.02 |
ENSG00000238727 |
. |
34372 |
0.23 |
| chr22_18049670_18050213 | 0.02 |
SLC25A18 |
solute carrier family 25 (glutamate carrier), member 18 |
6791 |
0.19 |
| chr18_21405936_21406114 | 0.02 |
LAMA3 |
laminin, alpha 3 |
7892 |
0.26 |
| chr5_154151043_154151438 | 0.02 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
14506 |
0.17 |
| chr12_20000310_20000490 | 0.02 |
ENSG00000200885 |
. |
123031 |
0.06 |
| chr8_110632793_110632963 | 0.02 |
SYBU |
syntabulin (syntaxin-interacting) |
12574 |
0.18 |
| chr12_123367754_123367930 | 0.02 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
6864 |
0.17 |
| chr22_24721245_24721396 | 0.02 |
SPECC1L |
sperm antigen with calponin homology and coiled-coil domains 1-like |
20956 |
0.18 |
| chr13_45145551_45145847 | 0.02 |
TSC22D1 |
TSC22 domain family, member 1 |
4693 |
0.31 |
| chr4_153703528_153703679 | 0.02 |
ARFIP1 |
ADP-ribosylation factor interacting protein 1 |
2466 |
0.28 |
| chr20_4669808_4669959 | 0.02 |
PRNP |
prion protein |
2694 |
0.3 |
| chr2_9139987_9140138 | 0.02 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
3732 |
0.3 |
| chr16_62068993_62069255 | 0.02 |
CDH8 |
cadherin 8, type 2 |
86 |
0.98 |
| chr2_44472177_44472328 | 0.02 |
ENSG00000239052 |
. |
5452 |
0.2 |
| chr8_96031017_96031168 | 0.02 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
6122 |
0.18 |
| chr18_52763849_52764148 | 0.02 |
ENSG00000252437 |
. |
49878 |
0.16 |
| chr15_57371657_57371950 | 0.02 |
ENSG00000222586 |
. |
113208 |
0.06 |
| chr17_79315981_79316442 | 0.02 |
TMEM105 |
transmembrane protein 105 |
11737 |
0.13 |
| chr3_71834358_71834666 | 0.02 |
PROK2 |
prokineticin 2 |
155 |
0.96 |
| chr8_19112471_19112622 | 0.02 |
SH2D4A |
SH2 domain containing 4A |
58582 |
0.16 |
| chr1_186808344_186808515 | 0.02 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
10307 |
0.31 |
| chr21_40386589_40386894 | 0.02 |
ENSG00000272015 |
. |
120032 |
0.05 |
| chr18_62105511_62105662 | 0.02 |
ENSG00000252818 |
. |
111156 |
0.07 |
| chr8_23656283_23656639 | 0.02 |
ENSG00000207027 |
. |
7429 |
0.2 |
| chr5_40224048_40224291 | 0.02 |
ENSG00000199361 |
. |
45491 |
0.19 |
| chrX_46442135_46442322 | 0.02 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
9009 |
0.23 |
| chr4_173063595_173063799 | 0.02 |
GALNTL6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
90101 |
0.11 |
| chr5_99606023_99606303 | 0.02 |
ENSG00000207077 |
. |
116787 |
0.07 |
| chr4_5052773_5052975 | 0.02 |
STK32B |
serine/threonine kinase 32B |
295 |
0.93 |
| chr6_76645073_76645368 | 0.02 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
454 |
0.81 |
| chr2_73463870_73464021 | 0.02 |
CCT7 |
chaperonin containing TCP1, subunit 7 (eta) |
2499 |
0.19 |
| chr1_153747814_153748820 | 0.02 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
549 |
0.6 |
| chr9_687058_687461 | 0.02 |
RP11-130C19.3 |
|
1704 |
0.41 |
| chr11_65256028_65256296 | 0.02 |
AP000769.1 |
Uncharacterized protein |
33434 |
0.08 |
| chr8_81933876_81934027 | 0.02 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
90352 |
0.09 |
| chr8_41363635_41363786 | 0.02 |
GOLGA7 |
golgin A7 |
15537 |
0.15 |
| chr16_73096665_73097349 | 0.02 |
ZFHX3 |
zinc finger homeobox 3 |
3410 |
0.3 |
| chr6_158254601_158254752 | 0.02 |
RP11-52J3.3 |
|
3071 |
0.24 |
| chr3_120626761_120627590 | 0.02 |
STXBP5L |
syntaxin binding protein 5-like |
14 |
0.99 |
| chr2_179314595_179315822 | 0.02 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
276 |
0.8 |
| chr21_38780543_38780694 | 0.02 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
10589 |
0.23 |
| chr3_138405652_138405803 | 0.02 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
27601 |
0.21 |
| chr17_2115742_2115893 | 0.02 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
214 |
0.85 |
| chr12_12008308_12008477 | 0.02 |
ETV6 |
ets variant 6 |
30479 |
0.24 |
| chr15_75335743_75336025 | 0.02 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
268 |
0.9 |
| chr17_46681017_46681197 | 0.02 |
HOXB6 |
homeobox B6 |
1227 |
0.18 |
| chr10_13380689_13380840 | 0.02 |
SEPHS1 |
selenophosphate synthetase 1 |
2026 |
0.31 |
| chr3_37031282_37031460 | 0.02 |
EPM2AIP1 |
EPM2A (laforin) interacting protein 1 |
3424 |
0.2 |
| chr8_1703441_1703788 | 0.02 |
CTD-2336O2.1 |
|
7849 |
0.21 |
| chr6_138866633_138866868 | 0.02 |
NHSL1 |
NHS-like 1 |
98 |
0.98 |
| chr5_122621760_122621911 | 0.02 |
CEP120 |
centrosomal protein 120kDa |
137162 |
0.05 |
| chr2_177447944_177448206 | 0.02 |
ENSG00000252027 |
. |
81329 |
0.1 |
| chr11_31825425_31825576 | 0.02 |
PAX6 |
paired box 6 |
2969 |
0.27 |
| chr3_177695433_177695584 | 0.02 |
ENSG00000199858 |
. |
30059 |
0.26 |
| chr4_41157110_41157449 | 0.02 |
ENSG00000207198 |
. |
41320 |
0.14 |
| chr14_77508251_77508402 | 0.02 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
13292 |
0.18 |
| chr22_40357384_40357560 | 0.02 |
RP3-370M22.8 |
|
1071 |
0.49 |
| chr4_184369530_184369754 | 0.02 |
CDKN2AIP |
CDKN2A interacting protein |
3792 |
0.23 |
| chr7_127857654_127857944 | 0.02 |
ENSG00000207705 |
. |
9875 |
0.19 |
| chr17_19373891_19374103 | 0.02 |
ENSG00000265335 |
. |
50405 |
0.08 |
| chr17_38480138_38480579 | 0.02 |
RARA |
retinoic acid receptor, alpha |
5821 |
0.12 |
| chr3_190436802_190436953 | 0.02 |
ENSG00000223117 |
. |
76925 |
0.11 |
| chr7_113722711_113723554 | 0.02 |
FOXP2 |
forkhead box P2 |
3483 |
0.3 |
| chr9_95166770_95166921 | 0.02 |
OGN |
osteoglycin |
4 |
0.98 |
| chr15_36145125_36145276 | 0.02 |
ENSG00000265098 |
. |
73857 |
0.13 |
| chr13_42137393_42137544 | 0.02 |
ENSG00000264190 |
. |
5063 |
0.28 |
| chr17_21154446_21154597 | 0.02 |
C17orf103 |
chromosome 17 open reading frame 103 |
2057 |
0.26 |
| chr5_138206284_138206563 | 0.02 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
3304 |
0.25 |
| chr7_79908056_79908207 | 0.02 |
ENSG00000244392 |
. |
32889 |
0.17 |
| chr3_157217019_157217987 | 0.02 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
58 |
0.98 |
| chr11_58975341_58975492 | 0.02 |
MPEG1 |
macrophage expressed 1 |
5008 |
0.18 |
| chr2_208602192_208602343 | 0.02 |
CCNYL1 |
cyclin Y-like 1 |
4731 |
0.16 |
| chr2_169402424_169402596 | 0.02 |
ENSG00000265694 |
. |
36943 |
0.17 |
| chr3_54157684_54157865 | 0.02 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
286 |
0.95 |
| chr1_184836333_184836484 | 0.02 |
ENSG00000252222 |
. |
45785 |
0.15 |
| chr6_137561305_137561456 | 0.02 |
IFNGR1 |
interferon gamma receptor 1 |
20794 |
0.24 |
| chr13_40683698_40683888 | 0.02 |
ENSG00000207458 |
. |
117171 |
0.07 |
| chr12_32701711_32701862 | 0.02 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
14528 |
0.25 |
| chr22_46472382_46472746 | 0.02 |
FLJ27365 |
hsa-mir-4763 |
3628 |
0.12 |
| chr6_38420811_38420962 | 0.02 |
BTBD9-AS1 |
BTBD9 antisense RNA 1 |
28582 |
0.22 |
| chr6_130005371_130005537 | 0.02 |
ARHGAP18 |
Rho GTPase activating protein 18 |
25916 |
0.23 |
| chr2_28523371_28523522 | 0.02 |
AC093690.1 |
|
9880 |
0.25 |
| chr14_29047076_29047227 | 0.02 |
RP11-966I7.1 |
|
187318 |
0.03 |
| chr3_16525949_16526100 | 0.02 |
RFTN1 |
raftlin, lipid raft linker 1 |
1652 |
0.49 |
| chr1_161187234_161187476 | 0.02 |
FCER1G |
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
2268 |
0.11 |
| chr18_55890673_55891004 | 0.02 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
2035 |
0.39 |
| chr3_4517627_4517778 | 0.02 |
SUMF1 |
sulfatase modifying factor 1 |
8737 |
0.2 |
| chr10_76092894_76093045 | 0.02 |
ADK |
adenosine kinase |
156448 |
0.04 |
| chr15_37878446_37878600 | 0.02 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
335617 |
0.01 |
| chr10_54077811_54078125 | 0.02 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
3912 |
0.28 |
| chr4_33926484_33926635 | 0.02 |
ENSG00000251755 |
. |
140002 |
0.05 |
| chr10_112410971_112411122 | 0.02 |
RBM20 |
RNA binding motif protein 20 |
6891 |
0.16 |
| chr3_181431793_181431944 | 0.02 |
SOX2 |
SRY (sex determining region Y)-box 2 |
2146 |
0.37 |
| chr4_154149984_154150203 | 0.02 |
TRIM2 |
tripartite motif containing 2 |
24475 |
0.17 |
| chr8_67624599_67625774 | 0.02 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
533 |
0.76 |
| chr9_21837665_21837816 | 0.02 |
MTAP |
methylthioadenosine phosphorylase |
16983 |
0.17 |
| chr1_81703498_81704139 | 0.01 |
ENSG00000223026 |
. |
13656 |
0.28 |
| chr10_106112880_106113082 | 0.01 |
CCDC147-AS1 |
CCDC147 antisense RNA 1 (head to head) |
352 |
0.61 |
| chr15_57509980_57510170 | 0.01 |
TCF12 |
transcription factor 12 |
1553 |
0.51 |
| chr1_28744223_28744374 | 0.01 |
ENSG00000206779 |
. |
4868 |
0.17 |
| chr3_59412823_59412974 | 0.01 |
C3orf67 |
chromosome 3 open reading frame 67 |
377088 |
0.01 |
| chr10_33530135_33530286 | 0.01 |
NRP1 |
neuropilin 1 |
22422 |
0.19 |
| chr6_11229774_11229925 | 0.01 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
3042 |
0.32 |
| chr12_95009320_95010015 | 0.01 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
170 |
0.97 |
| chr6_16200733_16201077 | 0.01 |
ENSG00000251831 |
. |
4340 |
0.21 |
| chr2_237878607_237878758 | 0.01 |
ENSG00000202341 |
. |
45266 |
0.17 |
| chr6_151409287_151409438 | 0.01 |
RP1-292B18.3 |
|
32169 |
0.16 |
| chr13_99930587_99930738 | 0.01 |
GPR18 |
G protein-coupled receptor 18 |
16664 |
0.18 |
| chr12_80322059_80322210 | 0.01 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
6361 |
0.21 |
| chr8_105655635_105655790 | 0.01 |
RP11-127H5.1 |
Uncharacterized protein |
52751 |
0.14 |
| chr15_43541860_43542154 | 0.01 |
ENSG00000202211 |
. |
2129 |
0.22 |
| chr8_129879414_129879565 | 0.01 |
ENSG00000221351 |
. |
47449 |
0.2 |
| chr1_56762365_56762516 | 0.01 |
ENSG00000223307 |
. |
80650 |
0.12 |
| chr2_54827541_54827692 | 0.01 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
42085 |
0.13 |
| chr7_101688653_101689251 | 0.01 |
CTA-357J21.1 |
|
12649 |
0.24 |
| chr4_82134488_82134639 | 0.01 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
1625 |
0.48 |
| chr6_106280114_106280265 | 0.01 |
ENSG00000200198 |
. |
72059 |
0.13 |
| chr7_121945437_121945668 | 0.01 |
FEZF1-AS1 |
FEZF1 antisense RNA 1 |
548 |
0.63 |
| chr7_2439740_2440019 | 0.01 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
3344 |
0.25 |
| chr13_30497549_30497716 | 0.01 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
3156 |
0.37 |
| chr10_65558718_65559094 | 0.01 |
REEP3 |
receptor accessory protein 3 |
277783 |
0.01 |
| chr14_35170020_35170171 | 0.01 |
CFL2 |
cofilin 2 (muscle) |
12939 |
0.18 |
| chr6_139866897_139867163 | 0.01 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
171273 |
0.04 |
| chr10_124254663_124255115 | 0.01 |
HTRA1 |
HtrA serine peptidase 1 |
11318 |
0.2 |
| chr17_37050262_37050413 | 0.01 |
RP1-56K13.2 |
|
10516 |
0.1 |
| chr1_232021274_232021425 | 0.01 |
DISC1-IT1 |
DISC1 intronic transcript 1 (non-protein coding) |
40231 |
0.21 |
| chr15_37075326_37075477 | 0.01 |
C15orf41 |
chromosome 15 open reading frame 41 |
27255 |
0.23 |
| chr5_58676265_58676426 | 0.01 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
23544 |
0.28 |
| chr15_67399699_67399853 | 0.01 |
SMAD3 |
SMAD family member 3 |
8759 |
0.25 |
| chr19_48219590_48219888 | 0.01 |
EHD2 |
EH-domain containing 2 |
3060 |
0.19 |
| chr12_16759185_16759513 | 0.01 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
116 |
0.98 |
| chr4_25238322_25238473 | 0.01 |
PI4K2B |
phosphatidylinositol 4-kinase type 2 beta |
2800 |
0.34 |
| chr3_14476500_14476651 | 0.01 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
2397 |
0.35 |
| chr11_134009594_134009775 | 0.01 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
28683 |
0.16 |
| chr4_16723232_16723461 | 0.01 |
LDB2 |
LIM domain binding 2 |
47410 |
0.2 |
| chr7_18794470_18794725 | 0.01 |
ENSG00000222164 |
. |
53305 |
0.17 |
| chr17_65430092_65430384 | 0.01 |
ENSG00000244610 |
. |
23168 |
0.12 |
| chr5_179757441_179757592 | 0.01 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
1132 |
0.52 |
| chr9_89891784_89891968 | 0.01 |
ENSG00000212421 |
. |
16511 |
0.28 |
| chr3_29800003_29800421 | 0.01 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
115912 |
0.07 |
| chr15_48959812_48960090 | 0.01 |
FBN1 |
fibrillin 1 |
21905 |
0.23 |
| chr11_128457327_128457522 | 0.01 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
29 |
0.98 |
| chr4_186704181_186704332 | 0.01 |
SORBS2 |
sorbin and SH3 domain containing 2 |
7190 |
0.26 |
| chr13_72402750_72402901 | 0.01 |
DACH1 |
dachshund homolog 1 (Drosophila) |
38082 |
0.23 |
| chrX_47056180_47056331 | 0.01 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
343 |
0.82 |
| chr14_57276490_57276707 | 0.01 |
OTX2 |
orthodenticle homeobox 2 |
568 |
0.71 |
| chr2_144855004_144855155 | 0.01 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
138225 |
0.05 |
| chr2_101840427_101840591 | 0.01 |
CNOT11 |
CCR4-NOT transcription complex, subunit 11 |
28755 |
0.16 |
| chr21_17882200_17882351 | 0.01 |
ENSG00000207638 |
. |
29134 |
0.19 |
| chr7_30357859_30358186 | 0.01 |
ENSG00000207771 |
. |
28612 |
0.17 |
| chr16_3333511_3334475 | 0.01 |
ZNF263 |
zinc finger protein 263 |
38 |
0.95 |
| chr15_99864560_99864718 | 0.01 |
AC022819.2 |
Uncharacterized protein |
4811 |
0.23 |
| chr12_91720658_91720809 | 0.01 |
DCN |
decorin |
143833 |
0.05 |
| chr3_149298860_149299373 | 0.01 |
WWTR1 |
WW domain containing transcription regulator 1 |
5077 |
0.23 |
| chr5_138060722_138060873 | 0.01 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
27989 |
0.15 |
| chr1_31500523_31500674 | 0.01 |
PUM1 |
pumilio RNA-binding family member 1 |
32647 |
0.16 |
| chr7_102083951_102084102 | 0.01 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
7280 |
0.09 |
| chr3_171024511_171024705 | 0.01 |
TNIK |
TRAF2 and NCK interacting kinase |
81108 |
0.1 |
| chr14_71609169_71609320 | 0.01 |
PCNX |
pecanex homolog (Drosophila) |
33507 |
0.21 |
| chr4_19852357_19852510 | 0.01 |
SLIT2 |
slit homolog 2 (Drosophila) |
402450 |
0.01 |
| chr18_53214040_53214242 | 0.01 |
TCF4 |
transcription factor 4 |
36141 |
0.18 |
| chr17_5692957_5693108 | 0.01 |
WSCD1 |
WSC domain containing 1 |
16786 |
0.27 |
| chr8_42123666_42123817 | 0.01 |
RP11-231D20.2 |
|
525 |
0.74 |
| chr2_13059353_13059504 | 0.01 |
ENSG00000264370 |
. |
181935 |
0.03 |
| chr9_129896025_129896176 | 0.01 |
ANGPTL2 |
angiopoietin-like 2 |
10938 |
0.23 |
| chr3_196352166_196352317 | 0.01 |
LINC01063 |
long intergenic non-protein coding RNA 1063 |
7217 |
0.14 |
| chr12_104973868_104974019 | 0.01 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
8683 |
0.24 |
| chr6_112542856_112543007 | 0.01 |
RP11-506B6.6 |
|
14363 |
0.17 |
| chr3_156423919_156424189 | 0.01 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
25099 |
0.2 |
| chr11_61159610_61160825 | 0.01 |
TMEM216 |
transmembrane protein 216 |
352 |
0.79 |
Network of associatons between targets according to the STRING database.