Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
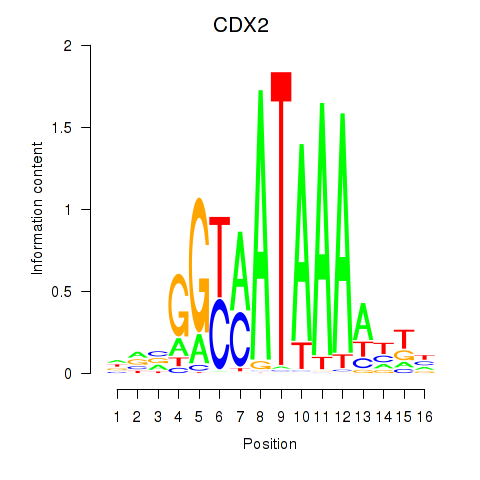
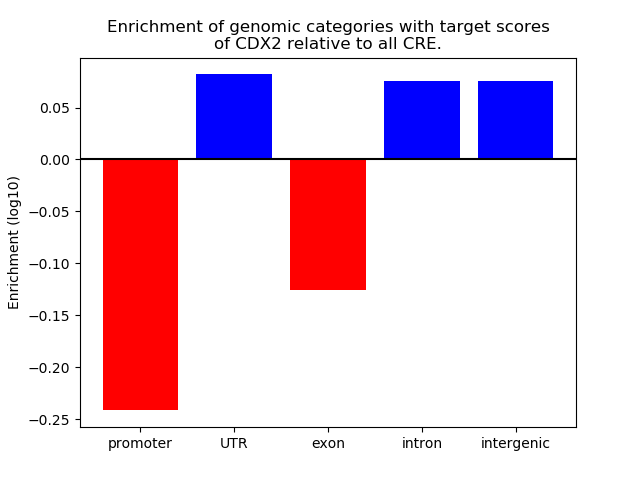
Results for CDX2
Z-value: 1.16
Transcription factors associated with CDX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX2
|
ENSG00000165556.9 | caudal type homeobox 2 |
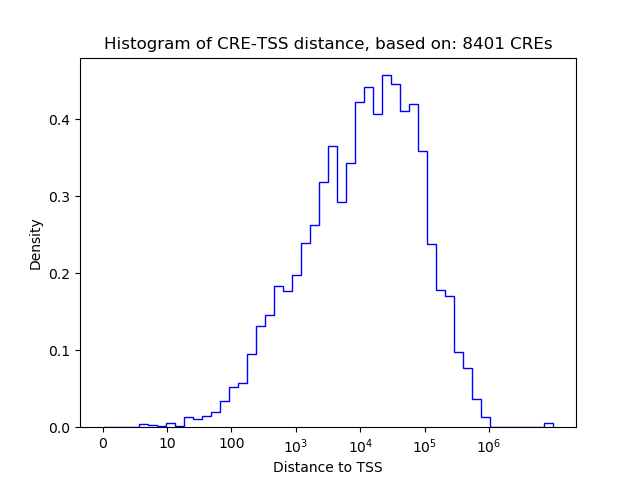
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_28542515_28542921 | CDX2 | 2558 | 0.211068 | -0.44 | 2.3e-01 | Click! |
| chr13_28552170_28552321 | CDX2 | 6969 | 0.146887 | -0.19 | 6.2e-01 | Click! |
| chr13_28534474_28535073 | CDX2 | 10503 | 0.135990 | -0.08 | 8.5e-01 | Click! |
| chr13_28535463_28535614 | CDX2 | 9738 | 0.137578 | -0.05 | 9.1e-01 | Click! |
| chr13_28534258_28534409 | CDX2 | 10943 | 0.135126 | 0.03 | 9.3e-01 | Click! |
Activity of the CDX2 motif across conditions
Conditions sorted by the z-value of the CDX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
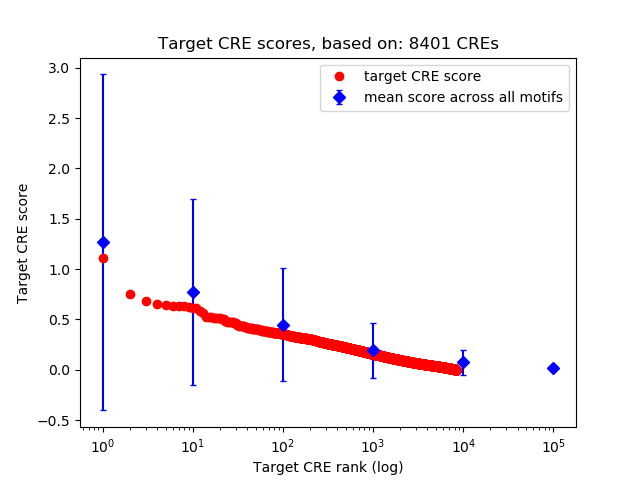
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_134464463_134465343 | 1.11 |
CALD1 |
caldesmon 1 |
474 |
0.89 |
| chr3_87036535_87037613 | 0.75 |
VGLL3 |
vestigial like 3 (Drosophila) |
2778 |
0.42 |
| chr11_63639674_63640171 | 0.69 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
3174 |
0.18 |
| chr11_132813237_132813595 | 0.66 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
150 |
0.98 |
| chr8_37350646_37351319 | 0.65 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr5_14036568_14037099 | 0.64 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
92181 |
0.09 |
| chr5_120114458_120114855 | 0.63 |
ENSG00000222609 |
. |
68121 |
0.14 |
| chr15_96883311_96883827 | 0.63 |
ENSG00000222651 |
. |
7079 |
0.16 |
| chr4_159731146_159731380 | 0.62 |
FNIP2 |
folliculin interacting protein 2 |
3863 |
0.25 |
| chr4_87516254_87516849 | 0.62 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
295 |
0.87 |
| chr6_53928552_53928746 | 0.61 |
MLIP-AS1 |
MLIP antisense RNA 1 |
15875 |
0.2 |
| chr10_74854370_74854615 | 0.59 |
P4HA1 |
prolyl 4-hydroxylase, alpha polypeptide I |
2116 |
0.19 |
| chr10_33620428_33620906 | 0.57 |
NRP1 |
neuropilin 1 |
2643 |
0.37 |
| chr18_9615856_9616007 | 0.53 |
PPP4R1 |
protein phosphatase 4, regulatory subunit 1 |
693 |
0.62 |
| chr12_7651228_7651481 | 0.52 |
CD163 |
CD163 molecule |
3979 |
0.25 |
| chr7_132299406_132299928 | 0.52 |
PLXNA4 |
plexin A4 |
33780 |
0.18 |
| chr19_4061458_4061934 | 0.52 |
CTD-2622I13.3 |
|
1051 |
0.37 |
| chr3_112358713_112358917 | 0.52 |
CCDC80 |
coiled-coil domain containing 80 |
1301 |
0.54 |
| chr1_221067632_221067991 | 0.52 |
HLX |
H2.0-like homeobox |
13227 |
0.2 |
| chr9_21964953_21966022 | 0.51 |
C9orf53 |
chromosome 9 open reading frame 53 |
1650 |
0.31 |
| chr15_39871853_39872515 | 0.51 |
THBS1 |
thrombospondin 1 |
1096 |
0.52 |
| chr7_116164164_116164509 | 0.50 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
503 |
0.76 |
| chr7_5467738_5468320 | 0.49 |
TNRC18 |
trinucleotide repeat containing 18 |
2984 |
0.19 |
| chr21_36167680_36167831 | 0.48 |
AP000330.8 |
|
49621 |
0.16 |
| chr17_69773494_69773789 | 0.48 |
AC007461.1 |
Uncharacterized protein |
262523 |
0.02 |
| chr13_36045367_36045518 | 0.48 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
5390 |
0.22 |
| chr1_231963429_231963641 | 0.48 |
DISC1-IT1 |
DISC1 intronic transcript 1 (non-protein coding) |
98045 |
0.08 |
| chr12_1465959_1466110 | 0.47 |
RP5-951N9.2 |
|
28965 |
0.18 |
| chr10_4285370_4285529 | 0.46 |
ENSG00000207124 |
. |
271695 |
0.02 |
| chr15_99788624_99789059 | 0.46 |
TTC23 |
tetratricopeptide repeat domain 23 |
895 |
0.51 |
| chr5_92921648_92921875 | 0.45 |
ENSG00000237187 |
. |
407 |
0.85 |
| chr10_114714851_114715858 | 0.44 |
RP11-57H14.2 |
|
3720 |
0.26 |
| chr2_45823201_45823510 | 0.44 |
SRBD1 |
S1 RNA binding domain 1 |
15065 |
0.21 |
| chr5_158611818_158612659 | 0.44 |
RNF145 |
ring finger protein 145 |
22404 |
0.16 |
| chr4_89977525_89978081 | 0.44 |
FAM13A |
family with sequence similarity 13, member A |
508 |
0.85 |
| chr1_224022481_224022658 | 0.43 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
11063 |
0.26 |
| chr15_48960228_48960461 | 0.43 |
FBN1 |
fibrillin 1 |
22298 |
0.23 |
| chr10_69916813_69916964 | 0.42 |
ENSG00000222371 |
. |
948 |
0.6 |
| chr17_45959589_45959832 | 0.42 |
SP2 |
Sp2 transcription factor |
13806 |
0.1 |
| chr13_76213824_76214088 | 0.42 |
LMO7 |
LIM domain 7 |
3497 |
0.21 |
| chr4_86700210_86700446 | 0.42 |
ARHGAP24 |
Rho GTPase activating protein 24 |
469 |
0.88 |
| chr19_41932700_41933275 | 0.42 |
B3GNT8 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
1648 |
0.18 |
| chr12_26379464_26379833 | 0.41 |
SSPN |
sarcospan |
31042 |
0.17 |
| chr3_29327656_29328192 | 0.41 |
RBMS3-AS3 |
RBMS3 antisense RNA 3 |
4293 |
0.26 |
| chr11_101918556_101918783 | 0.41 |
C11orf70 |
chromosome 11 open reading frame 70 |
480 |
0.77 |
| chr3_128207705_128209147 | 0.41 |
RP11-475N22.4 |
|
326 |
0.79 |
| chr1_65363518_65363760 | 0.41 |
JAK1 |
Janus kinase 1 |
68548 |
0.12 |
| chr13_73664940_73665091 | 0.41 |
ENSG00000200267 |
. |
22140 |
0.18 |
| chr8_65754459_65754681 | 0.41 |
CYP7B1 |
cytochrome P450, family 7, subfamily B, polypeptide 1 |
43252 |
0.2 |
| chr15_99789888_99790153 | 0.40 |
TTC23 |
tetratricopeptide repeat domain 23 |
205 |
0.9 |
| chr15_65805259_65805472 | 0.40 |
DPP8 |
dipeptidyl-peptidase 8 |
196 |
0.92 |
| chr4_114899439_114899761 | 0.40 |
ARSJ |
arylsulfatase family, member J |
552 |
0.84 |
| chr10_29923748_29924013 | 0.40 |
SVIL |
supervillin |
21 |
0.98 |
| chr4_157700078_157700308 | 0.40 |
RP11-154F14.2 |
|
62318 |
0.13 |
| chr10_117577472_117577686 | 0.40 |
GFRA1 |
GDNF family receptor alpha 1 |
437728 |
0.01 |
| chr8_58715313_58715521 | 0.40 |
FAM110B |
family with sequence similarity 110, member B |
191696 |
0.03 |
| chr7_123284039_123284287 | 0.40 |
LMOD2 |
leiomodin 2 (cardiac) |
11698 |
0.17 |
| chr2_197852125_197852688 | 0.39 |
ANKRD44 |
ankyrin repeat domain 44 |
12820 |
0.25 |
| chr6_105388737_105389249 | 0.39 |
LIN28B |
lin-28 homolog B (C. elegans) |
15930 |
0.23 |
| chr21_17489963_17490188 | 0.39 |
ENSG00000252273 |
. |
82246 |
0.11 |
| chr7_94029074_94029225 | 0.39 |
COL1A2 |
collagen, type I, alpha 2 |
5276 |
0.3 |
| chr7_114207737_114207888 | 0.39 |
ENSG00000202334 |
. |
46149 |
0.15 |
| chr7_113722711_113723554 | 0.39 |
FOXP2 |
forkhead box P2 |
3483 |
0.3 |
| chr18_21264816_21264967 | 0.38 |
LAMA3 |
laminin, alpha 3 |
4516 |
0.24 |
| chr14_23623374_23623574 | 0.38 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
128 |
0.92 |
| chr12_54427392_54427581 | 0.38 |
ENSG00000207571 |
. |
248 |
0.73 |
| chr19_45995836_45996758 | 0.38 |
RTN2 |
reticulon 2 |
228 |
0.87 |
| chr7_41740779_41741576 | 0.38 |
INHBA |
inhibin, beta A |
970 |
0.56 |
| chr7_38670068_38671048 | 0.38 |
AMPH |
amphiphysin |
462 |
0.87 |
| chr15_86164203_86164426 | 0.38 |
RP11-815J21.3 |
|
6606 |
0.15 |
| chr15_38547811_38548278 | 0.37 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
2662 |
0.42 |
| chr5_72733592_72733937 | 0.37 |
FOXD1 |
forkhead box D1 |
10588 |
0.21 |
| chr9_12859535_12860149 | 0.37 |
ENSG00000222658 |
. |
25478 |
0.2 |
| chr2_55239455_55239731 | 0.37 |
RTN4 |
reticulon 4 |
2008 |
0.3 |
| chr2_238596792_238597019 | 0.37 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
3883 |
0.27 |
| chr12_101188560_101188771 | 0.37 |
ANO4 |
anoctamin 4 |
70 |
0.98 |
| chr9_20558698_20558876 | 0.37 |
ENSG00000252324 |
. |
17429 |
0.2 |
| chr5_88238184_88238467 | 0.37 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
579 |
0.83 |
| chr5_98112819_98113202 | 0.37 |
RGMB |
repulsive guidance molecule family member b |
3671 |
0.26 |
| chr15_48436377_48436744 | 0.37 |
MYEF2 |
myelin expression factor 2 |
4963 |
0.17 |
| chr5_145319076_145319227 | 0.37 |
SH3RF2 |
SH3 domain containing ring finger 2 |
1711 |
0.46 |
| chr1_86846919_86847201 | 0.37 |
ODF2L |
outer dense fiber of sperm tails 2-like |
1819 |
0.42 |
| chr4_184509430_184509787 | 0.37 |
ENSG00000239116 |
. |
6154 |
0.2 |
| chr11_11605687_11605838 | 0.37 |
RP11-483L5.1 |
|
14131 |
0.22 |
| chrX_13906420_13906604 | 0.37 |
GPM6B |
glycoprotein M6B |
664 |
0.81 |
| chr4_85420275_85420452 | 0.36 |
NKX6-1 |
NK6 homeobox 1 |
760 |
0.77 |
| chr3_71158276_71158520 | 0.36 |
FOXP1 |
forkhead box P1 |
21346 |
0.28 |
| chr1_243962680_243962831 | 0.36 |
AKT3-IT1 |
AKT3 intronic transcript 1 (non-protein coding) |
5053 |
0.27 |
| chr6_114182367_114182913 | 0.36 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
4099 |
0.23 |
| chr13_65686743_65686894 | 0.36 |
ENSG00000221685 |
. |
751487 |
0.0 |
| chr13_21654404_21654812 | 0.36 |
LATS2 |
large tumor suppressor kinase 2 |
18922 |
0.14 |
| chr10_117577799_117577950 | 0.36 |
GFRA1 |
GDNF family receptor alpha 1 |
437433 |
0.01 |
| chr13_80912572_80913077 | 0.36 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
970 |
0.67 |
| chr4_27219934_27220157 | 0.36 |
ENSG00000222206 |
. |
5004 |
0.36 |
| chr18_25757173_25757741 | 0.36 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
47 |
0.99 |
| chr5_124748629_124749002 | 0.36 |
ENSG00000222107 |
. |
61991 |
0.16 |
| chr3_30730881_30731077 | 0.36 |
RP11-1024P17.1 |
|
8174 |
0.3 |
| chr5_88174722_88174873 | 0.36 |
MEF2C |
myocyte enhancer factor 2C |
1003 |
0.61 |
| chr10_112295759_112296296 | 0.36 |
SMC3 |
structural maintenance of chromosomes 3 |
31422 |
0.13 |
| chr12_117036919_117037510 | 0.36 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
23558 |
0.24 |
| chr18_74189277_74189488 | 0.36 |
ZNF516 |
zinc finger protein 516 |
13353 |
0.17 |
| chr5_111090218_111090459 | 0.36 |
NREP |
neuronal regeneration related protein |
1610 |
0.42 |
| chr12_1428214_1428403 | 0.36 |
RP5-951N9.2 |
|
66691 |
0.11 |
| chr11_5642308_5642558 | 0.35 |
TRIM34 |
tripartite motif containing 34 |
1439 |
0.24 |
| chr7_116441355_116441506 | 0.35 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
9694 |
0.21 |
| chr17_74706029_74706760 | 0.35 |
MXRA7 |
matrix-remodelling associated 7 |
643 |
0.49 |
| chr6_148845639_148845819 | 0.35 |
ENSG00000223322 |
. |
353 |
0.93 |
| chr7_87987383_87987655 | 0.35 |
STEAP4 |
STEAP family member 4 |
51313 |
0.14 |
| chr4_152148275_152148426 | 0.35 |
SH3D19 |
SH3 domain containing 19 |
600 |
0.77 |
| chrX_34673275_34673426 | 0.35 |
TMEM47 |
transmembrane protein 47 |
2055 |
0.5 |
| chr5_16889789_16890008 | 0.35 |
MYO10 |
myosin X |
26738 |
0.18 |
| chr7_42147351_42147643 | 0.35 |
GLI3 |
GLI family zinc finger 3 |
119823 |
0.07 |
| chr5_112582414_112582565 | 0.35 |
MCC |
mutated in colorectal cancers |
12115 |
0.27 |
| chr9_80278787_80279042 | 0.35 |
GNA14 |
guanine nucleotide binding protein (G protein), alpha 14 |
15691 |
0.24 |
| chr16_15952010_15952161 | 0.34 |
MYH11 |
myosin, heavy chain 11, smooth muscle |
1195 |
0.43 |
| chr13_54519922_54520073 | 0.34 |
ENSG00000220990 |
. |
577906 |
0.0 |
| chr1_225839301_225840394 | 0.34 |
ENAH |
enabled homolog (Drosophila) |
571 |
0.78 |
| chr2_200604849_200605000 | 0.34 |
FTCDNL1 |
formiminotransferase cyclodeaminase N-terminal like |
110672 |
0.07 |
| chr2_65890354_65890505 | 0.34 |
ENSG00000265899 |
. |
3961 |
0.34 |
| chr20_47408037_47408263 | 0.34 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
36270 |
0.19 |
| chr14_30664453_30664650 | 0.34 |
PRKD1 |
protein kinase D1 |
3447 |
0.39 |
| chr20_49137388_49137672 | 0.34 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
10610 |
0.16 |
| chr6_121756849_121757212 | 0.34 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
192 |
0.94 |
| chr11_40311822_40311973 | 0.33 |
LRRC4C |
leucine rich repeat containing 4C |
2783 |
0.39 |
| chr20_57604835_57604986 | 0.33 |
ATP5E |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
2437 |
0.22 |
| chr21_19157038_19157189 | 0.33 |
AL109761.5 |
|
8692 |
0.24 |
| chr14_75478720_75478902 | 0.33 |
RP11-950C14.3 |
|
3627 |
0.15 |
| chr6_165504753_165505087 | 0.33 |
C6orf118 |
chromosome 6 open reading frame 118 |
217422 |
0.02 |
| chrX_13007277_13007565 | 0.33 |
TMSB4X |
thymosin beta 4, X-linked |
13644 |
0.22 |
| chr15_92722178_92722335 | 0.33 |
RP11-24J19.1 |
|
6590 |
0.32 |
| chr19_52452690_52452938 | 0.33 |
HCCAT3 |
hepatocellular carcinoma associated transcript 3 (non-protein coding) |
399 |
0.77 |
| chr1_78892388_78892539 | 0.33 |
ENSG00000212308 |
. |
51510 |
0.15 |
| chr16_65104457_65104608 | 0.33 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
1584 |
0.58 |
| chr1_179111271_179111703 | 0.33 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
692 |
0.72 |
| chr1_59859347_59859498 | 0.33 |
FGGY |
FGGY carbohydrate kinase domain containing |
83663 |
0.11 |
| chr5_128796390_128797012 | 0.33 |
ADAMTS19-AS1 |
ADAMTS19 antisense RNA 1 |
319 |
0.77 |
| chr10_33274227_33274378 | 0.33 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
7089 |
0.27 |
| chr18_74200108_74200360 | 0.33 |
ZNF516 |
zinc finger protein 516 |
2501 |
0.24 |
| chr2_64515395_64515553 | 0.33 |
ENSG00000264297 |
. |
52419 |
0.14 |
| chr5_59893443_59893927 | 0.33 |
DEPDC1B |
DEP domain containing 1B |
102276 |
0.07 |
| chr1_170631765_170631991 | 0.33 |
PRRX1 |
paired related homeobox 1 |
397 |
0.91 |
| chr9_12775321_12775833 | 0.33 |
LURAP1L |
leucine rich adaptor protein 1-like |
557 |
0.78 |
| chr7_3476765_3477156 | 0.33 |
ENSG00000211544 |
. |
18261 |
0.27 |
| chr2_190244069_190244254 | 0.33 |
WDR75 |
WD repeat domain 75 |
61998 |
0.12 |
| chr2_200317783_200318056 | 0.33 |
SATB2 |
SATB homeobox 2 |
2892 |
0.31 |
| chr2_208549036_208549250 | 0.33 |
ENSG00000200764 |
. |
3541 |
0.18 |
| chr11_65058660_65059302 | 0.32 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
9003 |
0.12 |
| chr13_35962874_35963128 | 0.32 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
87831 |
0.09 |
| chr3_89350492_89350643 | 0.32 |
EPHA3 |
EPH receptor A3 |
193741 |
0.03 |
| chrX_47056761_47057047 | 0.32 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
306 |
0.84 |
| chr8_123200121_123200319 | 0.32 |
ENSG00000238901 |
. |
483310 |
0.01 |
| chr4_175075425_175075781 | 0.32 |
FBXO8 |
F-box protein 8 |
129211 |
0.05 |
| chr2_175192578_175192729 | 0.32 |
SP9 |
Sp9 transcription factor |
7021 |
0.17 |
| chr2_189840264_189840425 | 0.32 |
COL3A1 |
collagen, type III, alpha 1 |
1245 |
0.47 |
| chr1_94789725_94789884 | 0.32 |
ARHGAP29 |
Rho GTPase activating protein 29 |
86615 |
0.08 |
| chr2_181056671_181056823 | 0.32 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
184907 |
0.03 |
| chr2_201270113_201270684 | 0.32 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
12598 |
0.18 |
| chr12_27496070_27496221 | 0.32 |
ARNTL2 |
aryl hydrocarbon receptor nuclear translocator-like 2 |
10157 |
0.24 |
| chr4_157870413_157870730 | 0.32 |
PDGFC |
platelet derived growth factor C |
21484 |
0.21 |
| chr6_45523951_45524153 | 0.32 |
ENSG00000252738 |
. |
89789 |
0.09 |
| chr9_37574334_37574485 | 0.32 |
FBXO10 |
F-box protein 10 |
1841 |
0.32 |
| chr10_95241473_95241734 | 0.32 |
MYOF |
myoferlin |
348 |
0.88 |
| chr5_35851300_35851523 | 0.32 |
IL7R |
interleukin 7 receptor |
1386 |
0.45 |
| chr8_142141679_142142152 | 0.32 |
RP11-809O17.1 |
|
1855 |
0.33 |
| chr12_59311038_59311457 | 0.32 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
2080 |
0.36 |
| chr3_69494490_69494658 | 0.32 |
FRMD4B |
FERM domain containing 4B |
59144 |
0.16 |
| chr3_99761156_99761307 | 0.31 |
FILIP1L |
filamin A interacting protein 1-like |
72126 |
0.09 |
| chr7_27169269_27169568 | 0.31 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
178 |
0.67 |
| chr3_59101471_59101696 | 0.31 |
C3orf67 |
chromosome 3 open reading frame 67 |
65773 |
0.15 |
| chr22_28538513_28538664 | 0.31 |
TTC28 |
tetratricopeptide repeat domain 28 |
48465 |
0.15 |
| chr5_96895914_96896065 | 0.31 |
CTD-2215E18.2 |
|
358405 |
0.01 |
| chr7_89058204_89058355 | 0.31 |
ENSG00000239075 |
. |
325761 |
0.01 |
| chr4_138450639_138450790 | 0.31 |
PCDH18 |
protocadherin 18 |
2851 |
0.43 |
| chr6_56235163_56235375 | 0.31 |
COL21A1 |
collagen, type XXI, alpha 1 |
23623 |
0.2 |
| chr4_30865130_30865281 | 0.31 |
RP11-619J20.1 |
|
69613 |
0.14 |
| chr4_48024272_48024423 | 0.31 |
NIPAL1 |
NIPA-like domain containing 1 |
2797 |
0.23 |
| chr7_27646157_27646308 | 0.31 |
HIBADH |
3-hydroxyisobutyrate dehydrogenase |
41143 |
0.19 |
| chr6_42537305_42537764 | 0.31 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
5734 |
0.24 |
| chr6_82707021_82707202 | 0.31 |
ENSG00000223044 |
. |
213046 |
0.02 |
| chr9_112543013_112543264 | 0.31 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
369 |
0.36 |
| chr6_158461752_158461952 | 0.31 |
SYNJ2 |
synaptojanin 2 |
23355 |
0.18 |
| chr5_123252700_123253101 | 0.31 |
CSNK1G3 |
casein kinase 1, gamma 3 |
328781 |
0.01 |
| chr7_116909406_116909564 | 0.31 |
AC002465.2 |
|
31430 |
0.19 |
| chr4_123588404_123588555 | 0.31 |
IL21 |
interleukin 21 |
46255 |
0.14 |
| chr3_57042329_57042482 | 0.31 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
31002 |
0.17 |
| chr3_98625382_98625613 | 0.31 |
ENSG00000207331 |
. |
1806 |
0.32 |
| chr5_139913473_139913662 | 0.31 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
3407 |
0.11 |
| chr13_24145710_24146057 | 0.31 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1080 |
0.66 |
| chr3_98618999_98619299 | 0.31 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
866 |
0.54 |
| chr13_32859694_32859925 | 0.31 |
FRY |
furry homolog (Drosophila) |
2179 |
0.28 |
| chr3_145879077_145879748 | 0.31 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
458 |
0.86 |
| chr3_179168052_179168298 | 0.31 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
758 |
0.66 |
| chr17_75539020_75539171 | 0.31 |
SEPT9 |
septin 9 |
61100 |
0.11 |
| chr2_14633353_14633613 | 0.31 |
FAM84A |
family with sequence similarity 84, member A |
139327 |
0.05 |
| chr5_54456050_54456451 | 0.31 |
GPX8 |
glutathione peroxidase 8 (putative) |
252 |
0.86 |
| chr10_101768167_101768400 | 0.31 |
DNMBP |
dynamin binding protein |
1393 |
0.42 |
| chr12_66122461_66123138 | 0.31 |
HMGA2 |
high mobility group AT-hook 2 |
95112 |
0.08 |
| chr22_45865037_45865472 | 0.30 |
RP1-102D24.5 |
|
20630 |
0.17 |
| chr11_89113858_89114276 | 0.30 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr8_60213383_60213534 | 0.30 |
ENSG00000201763 |
. |
154986 |
0.04 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 0.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.5 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.3 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.3 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.4 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.3 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.2 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.2 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.4 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.2 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.3 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 0.5 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.1 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.2 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.6 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.3 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.5 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.3 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0052257 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0032825 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.6 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.2 | GO:0043206 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.2 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.4 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.0 | GO:0072194 | positive regulation of smooth muscle cell differentiation(GO:0051152) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.0 | GO:2000696 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.3 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.6 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.0 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.2 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:1902692 | regulation of neuroblast proliferation(GO:1902692) |
| 0.0 | 0.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0060579 | ventral spinal cord interneuron differentiation(GO:0021514) ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0035850 | epithelial cell differentiation involved in kidney development(GO:0035850) |
| 0.0 | 0.1 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.0 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.0 | GO:0051883 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0048641 | regulation of skeletal muscle tissue development(GO:0048641) |
| 0.0 | 0.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 1.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:2000053 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.0 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:2000649 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0050951 | sensory perception of temperature stimulus(GO:0050951) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.6 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.1 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.5 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0030354 | melanin-concentrating hormone activity(GO:0030354) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.0 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.0 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.0 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |