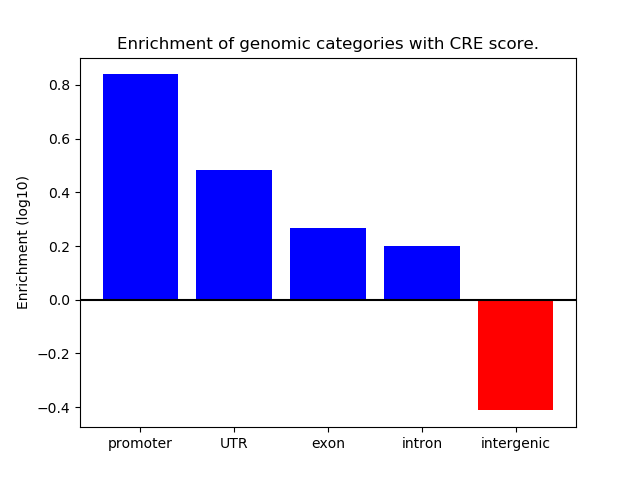
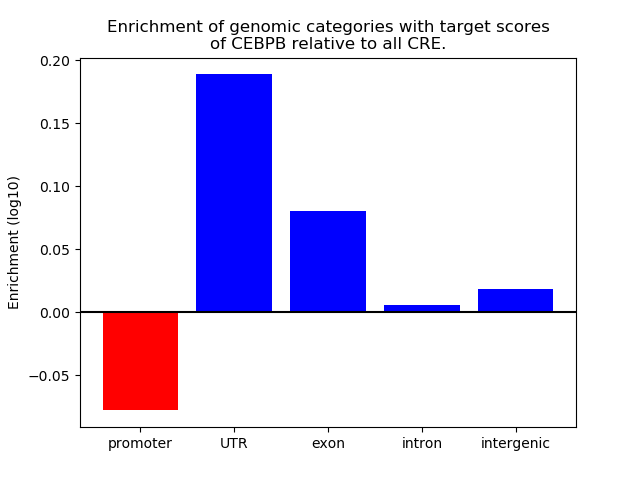
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
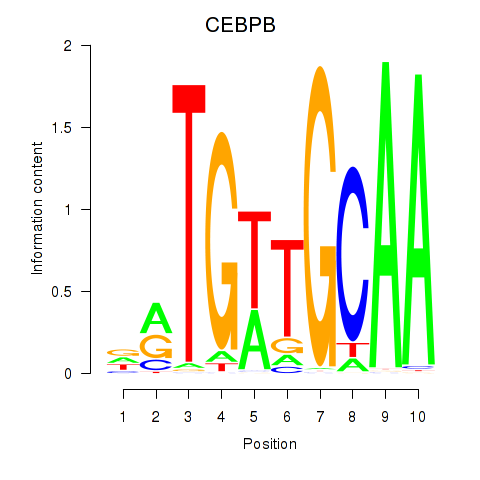
Results for CEBPB
Z-value: 1.89
Transcription factors associated with CEBPB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPB
|
ENSG00000172216.4 | CCAAT enhancer binding protein beta |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_48893065_48893216 | CEBPB | 85764 | 0.078934 | 0.86 | 3.2e-03 | Click! |
| chr20_48890286_48890437 | CEBPB | 82985 | 0.082311 | 0.82 | 7.3e-03 | Click! |
| chr20_48843063_48843214 | CEBPB | 35762 | 0.164960 | 0.80 | 8.9e-03 | Click! |
| chr20_48913935_48914086 | CEBPB | 106634 | 0.058344 | 0.78 | 1.4e-02 | Click! |
| chr20_48887644_48887838 | CEBPB | 80365 | 0.085644 | 0.76 | 1.7e-02 | Click! |
Activity of the CEBPB motif across conditions
Conditions sorted by the z-value of the CEBPB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
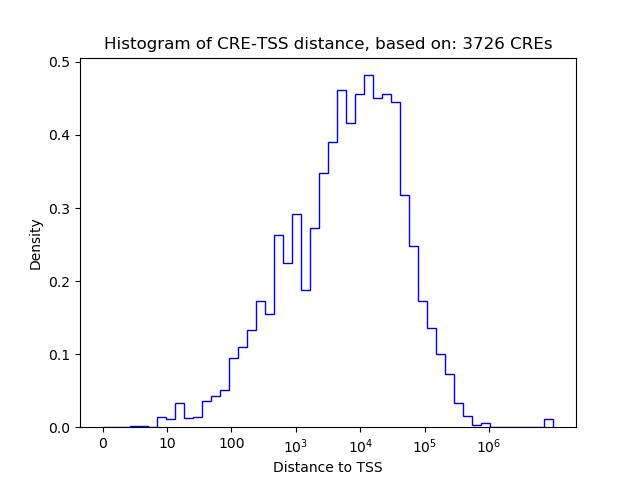
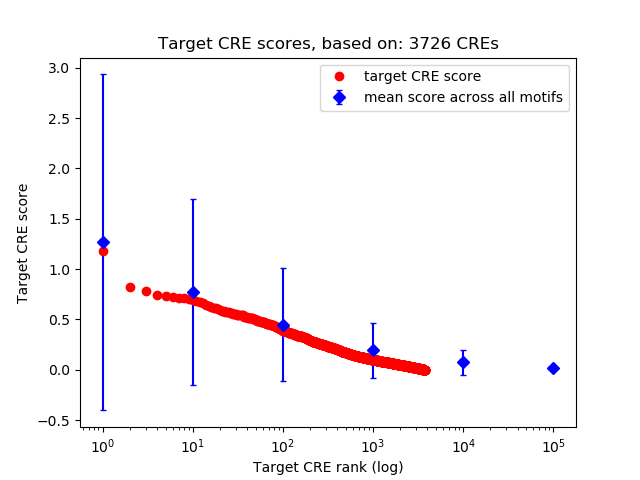
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_104827364_104827515 | 1.18 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
14 |
0.98 |
| chr9_132650035_132650502 | 0.82 |
FNBP1 |
formin binding protein 1 |
31321 |
0.13 |
| chr1_89339742_89339893 | 0.78 |
GTF2B |
general transcription factor IIB |
17362 |
0.19 |
| chr13_31312055_31312206 | 0.75 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
2485 |
0.4 |
| chr17_56365676_56365827 | 0.73 |
MPO |
myeloperoxidase |
7455 |
0.13 |
| chr22_17588053_17588350 | 0.72 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
13942 |
0.14 |
| chr1_223310789_223310940 | 0.71 |
TLR5 |
toll-like receptor 5 |
235 |
0.96 |
| chr12_54693878_54694144 | 0.71 |
NFE2 |
nuclear factor, erythroid 2 |
788 |
0.32 |
| chr9_132106265_132106416 | 0.70 |
ENSG00000242281 |
. |
26400 |
0.19 |
| chr8_37739073_37739224 | 0.70 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
3513 |
0.18 |
| chr7_33089731_33090334 | 0.69 |
ENSG00000241420 |
. |
4330 |
0.18 |
| chr9_93564472_93565060 | 0.67 |
SYK |
spleen tyrosine kinase |
557 |
0.87 |
| chr15_94406450_94406737 | 0.67 |
ENSG00000222409 |
. |
33663 |
0.25 |
| chr19_43097999_43098150 | 0.64 |
CEACAM8 |
carcinoembryonic antigen-related cell adhesion molecule 8 |
1008 |
0.56 |
| chr6_34240254_34240462 | 0.63 |
C6orf1 |
chromosome 6 open reading frame 1 |
23111 |
0.18 |
| chr21_35889691_35889842 | 0.62 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
5193 |
0.21 |
| chr21_43615613_43615764 | 0.61 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
4111 |
0.23 |
| chr15_101782791_101782964 | 0.61 |
CHSY1 |
chondroitin sulfate synthase 1 |
9260 |
0.18 |
| chr4_121736506_121736657 | 0.60 |
ENSG00000212359 |
. |
4957 |
0.33 |
| chr12_54893043_54893194 | 0.60 |
NCKAP1L |
NCK-associated protein 1-like |
550 |
0.7 |
| chr4_26275263_26275414 | 0.59 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
1109 |
0.68 |
| chr1_212781685_212782134 | 0.59 |
ATF3 |
activating transcription factor 3 |
103 |
0.96 |
| chr1_167199348_167199534 | 0.58 |
POU2F1 |
POU class 2 homeobox 1 |
9298 |
0.18 |
| chr11_75473950_75474101 | 0.57 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
3468 |
0.13 |
| chr4_36312312_36312717 | 0.57 |
DTHD1 |
death domain containing 1 |
26870 |
0.2 |
| chr19_54327344_54327551 | 0.56 |
NLRP12 |
NLR family, pyrin domain containing 12 |
124 |
0.86 |
| chr6_106970433_106970694 | 0.56 |
AIM1 |
absent in melanoma 1 |
10833 |
0.2 |
| chr2_64447057_64447208 | 0.56 |
AC074289.1 |
|
7863 |
0.27 |
| chr6_106898099_106898250 | 0.56 |
ENSG00000202386 |
. |
918 |
0.54 |
| chr4_40202894_40203061 | 0.55 |
RHOH |
ras homolog family member H |
1013 |
0.59 |
| chr17_8323121_8323272 | 0.55 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
6747 |
0.12 |
| chr12_54701075_54701275 | 0.55 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
6189 |
0.08 |
| chr11_94989698_94989849 | 0.55 |
RP11-712B9.2 |
|
24038 |
0.21 |
| chr3_137854795_137854946 | 0.54 |
A4GNT |
alpha-1,4-N-acetylglucosaminyltransferase |
3641 |
0.23 |
| chr7_37425762_37425913 | 0.54 |
ELMO1 |
engulfment and cell motility 1 |
32565 |
0.15 |
| chr15_80282157_80282308 | 0.54 |
BCL2A1 |
BCL2-related protein A1 |
18444 |
0.18 |
| chr13_98039572_98039723 | 0.53 |
ENSG00000202290 |
. |
24292 |
0.19 |
| chr1_112198825_112198976 | 0.53 |
ENSG00000201028 |
. |
5741 |
0.2 |
| chr16_84421_84572 | 0.52 |
WASIR2 |
WASH and IL9R antisense RNA 2 |
11586 |
0.1 |
| chr1_38311557_38311777 | 0.52 |
MTF1 |
metal-regulatory transcription factor 1 |
13625 |
0.09 |
| chr8_52782524_52782885 | 0.52 |
PCMTD1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
24399 |
0.15 |
| chr1_92057104_92057255 | 0.52 |
CDC7 |
cell division cycle 7 |
90484 |
0.09 |
| chr1_89737700_89738180 | 0.51 |
GBP5 |
guanylate binding protein 5 |
547 |
0.77 |
| chr22_18260050_18260218 | 0.51 |
BID |
BH3 interacting domain death agonist |
2703 |
0.26 |
| chr16_85793045_85793254 | 0.51 |
C16orf74 |
chromosome 16 open reading frame 74 |
8414 |
0.11 |
| chr12_51715569_51715862 | 0.51 |
BIN2 |
bridging integrator 2 |
2184 |
0.26 |
| chr8_37731641_37731792 | 0.51 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
3919 |
0.18 |
| chr20_19409135_19409286 | 0.51 |
ENSG00000221748 |
. |
87270 |
0.09 |
| chr10_3825891_3827255 | 0.50 |
KLF6 |
Kruppel-like factor 6 |
812 |
0.68 |
| chr3_69246175_69246326 | 0.49 |
FRMD4B |
FERM domain containing 4B |
2171 |
0.39 |
| chr9_36144726_36145049 | 0.49 |
GLIPR2 |
GLI pathogenesis-related 2 |
8145 |
0.19 |
| chr17_73308701_73308852 | 0.48 |
GRB2 |
growth factor receptor-bound protein 2 |
9058 |
0.1 |
| chr4_38787577_38787728 | 0.48 |
TLR10 |
toll-like receptor 10 |
3042 |
0.21 |
| chr2_101944298_101944469 | 0.48 |
ENSG00000264857 |
. |
18471 |
0.16 |
| chr13_28006863_28007014 | 0.48 |
GTF3A |
general transcription factor IIIA |
2843 |
0.23 |
| chr6_138132955_138133132 | 0.48 |
RP11-356I2.4 |
|
13988 |
0.21 |
| chr2_74197024_74197175 | 0.48 |
ENSG00000201876 |
. |
1766 |
0.3 |
| chr3_34198432_34198583 | 0.48 |
PDCD6IP |
programmed cell death 6 interacting protein |
358392 |
0.01 |
| chr9_139267151_139267437 | 0.48 |
CARD9 |
caspase recruitment domain family, member 9 |
839 |
0.45 |
| chr2_71680397_71681534 | 0.47 |
DYSF |
dysferlin |
113 |
0.98 |
| chr2_113870439_113870728 | 0.47 |
IL1RN |
interleukin 1 receptor antagonist |
1890 |
0.26 |
| chr13_115091452_115091603 | 0.47 |
CHAMP1 |
chromosome alignment maintaining phosphoprotein 1 |
4926 |
0.2 |
| chr10_14614242_14614420 | 0.47 |
FAM107B |
family with sequence similarity 107, member B |
7 |
0.98 |
| chr17_10018615_10018977 | 0.47 |
GAS7 |
growth arrest-specific 7 |
926 |
0.61 |
| chr5_114820222_114820373 | 0.46 |
FEM1C |
fem-1 homolog c (C. elegans) |
60294 |
0.12 |
| chr17_45743155_45743306 | 0.46 |
KPNB1 |
karyopherin (importin) beta 1 |
2501 |
0.2 |
| chr5_69770149_69770300 | 0.46 |
RP11-497H16.7 |
|
949 |
0.7 |
| chr5_171560548_171560699 | 0.45 |
ENSG00000266671 |
. |
52643 |
0.12 |
| chr9_130217644_130218078 | 0.45 |
LRSAM1 |
leucine rich repeat and sterile alpha motif containing 1 |
3327 |
0.15 |
| chr5_68915008_68915159 | 0.45 |
RP11-848G14.2 |
|
943 |
0.58 |
| chr7_112062054_112062259 | 0.45 |
IFRD1 |
interferon-related developmental regulator 1 |
867 |
0.64 |
| chr1_110415998_110416149 | 0.45 |
RP11-195M16.1 |
|
12813 |
0.17 |
| chr5_70423555_70423706 | 0.45 |
ENSG00000238451 |
. |
39171 |
0.18 |
| chr5_149791833_149792018 | 0.45 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
370 |
0.85 |
| chr5_70298657_70298808 | 0.45 |
NAIP |
NLR family, apoptosis inhibitory protein |
17805 |
0.22 |
| chr2_200603876_200604217 | 0.44 |
FTCDNL1 |
formiminotransferase cyclodeaminase N-terminal like |
111550 |
0.07 |
| chr10_28633323_28633474 | 0.44 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
9983 |
0.22 |
| chr10_134407309_134407460 | 0.44 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
14046 |
0.24 |
| chr17_33850543_33850694 | 0.44 |
RP11-1094M14.5 |
|
13329 |
0.11 |
| chr1_203273901_203274091 | 0.44 |
BTG2 |
BTG family, member 2 |
668 |
0.68 |
| chr5_69423238_69423389 | 0.44 |
ENSG00000265577 |
. |
52640 |
0.15 |
| chr14_88461258_88461409 | 0.43 |
GALC |
galactosylceramidase |
1324 |
0.33 |
| chr19_8634583_8634824 | 0.43 |
MYO1F |
myosin IF |
7619 |
0.12 |
| chr6_144474026_144474323 | 0.43 |
STX11 |
syntaxin 11 |
2511 |
0.36 |
| chr15_80262648_80263745 | 0.43 |
BCL2A1 |
BCL2-related protein A1 |
315 |
0.89 |
| chr12_9911079_9911493 | 0.42 |
CD69 |
CD69 molecule |
2211 |
0.26 |
| chr5_118609234_118609422 | 0.42 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
4879 |
0.21 |
| chr2_233941000_233941299 | 0.42 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
15960 |
0.18 |
| chr2_160418591_160418742 | 0.42 |
ENSG00000207117 |
. |
25809 |
0.19 |
| chr17_29645268_29645505 | 0.41 |
CTD-2370N5.3 |
|
272 |
0.86 |
| chr2_159070755_159070906 | 0.41 |
CCDC148-AS1 |
CCDC148 antisense RNA 1 |
47668 |
0.16 |
| chr5_158639058_158639320 | 0.41 |
RNF145 |
ring finger protein 145 |
2128 |
0.25 |
| chr18_54111502_54111653 | 0.41 |
TXNL1 |
thioredoxin-like 1 |
170145 |
0.04 |
| chr10_109449257_109449408 | 0.41 |
ENSG00000200079 |
. |
228014 |
0.02 |
| chr5_76152476_76152645 | 0.41 |
S100Z |
S100 calcium binding protein Z |
6636 |
0.18 |
| chr9_79230110_79230472 | 0.40 |
PRUNE2 |
prune homolog 2 (Drosophila) |
37174 |
0.16 |
| chr2_209048796_209048947 | 0.39 |
C2orf80 |
chromosome 2 open reading frame 80 |
1123 |
0.39 |
| chr7_37009137_37009288 | 0.39 |
ELMO1 |
engulfment and cell motility 1 |
15453 |
0.2 |
| chr17_68166405_68166607 | 0.39 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
830 |
0.45 |
| chr11_118111552_118111703 | 0.39 |
MPZL3 |
myelin protein zero-like 3 |
11435 |
0.13 |
| chr11_44991792_44991943 | 0.39 |
TP53I11 |
tumor protein p53 inducible protein 11 |
19259 |
0.24 |
| chrX_39610263_39610414 | 0.39 |
ENSG00000264618 |
. |
35739 |
0.21 |
| chr4_38857928_38858431 | 0.38 |
TLR6 |
toll-like receptor 6 |
258 |
0.48 |
| chr6_41169275_41169428 | 0.38 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
419 |
0.77 |
| chr3_169762562_169762818 | 0.38 |
GPR160 |
G protein-coupled receptor 160 |
4953 |
0.16 |
| chr9_123922153_123922304 | 0.38 |
CNTRL |
centriolin |
2107 |
0.3 |
| chr6_116950636_116950920 | 0.38 |
RSPH4A |
radial spoke head 4 homolog A (Chlamydomonas) |
12991 |
0.16 |
| chr2_38871082_38871242 | 0.38 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
21890 |
0.15 |
| chr20_58630648_58631293 | 0.38 |
C20orf197 |
chromosome 20 open reading frame 197 |
10 |
0.98 |
| chr2_169928702_169928853 | 0.38 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
233 |
0.94 |
| chr3_10231782_10231933 | 0.38 |
IRAK2 |
interleukin-1 receptor-associated kinase 2 |
25308 |
0.1 |
| chr15_55695695_55695873 | 0.38 |
CCPG1 |
cell cycle progression 1 |
4466 |
0.19 |
| chr3_128374531_128374682 | 0.38 |
RPN1 |
ribophorin I |
4914 |
0.23 |
| chr20_10485640_10485791 | 0.38 |
SLX4IP |
SLX4 interacting protein |
69764 |
0.11 |
| chr11_46025287_46025438 | 0.37 |
PHF21A |
PHD finger protein 21A |
57942 |
0.1 |
| chr19_43032769_43032982 | 0.37 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
214 |
0.93 |
| chr1_153337646_153337797 | 0.37 |
S100A9 |
S100 calcium binding protein A9 |
7391 |
0.11 |
| chr11_64114770_64114921 | 0.37 |
CCDC88B |
coiled-coil domain containing 88B |
1458 |
0.2 |
| chr12_69181196_69181347 | 0.36 |
AC124890.1 |
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
4854 |
0.17 |
| chr15_66662520_66662887 | 0.36 |
TIPIN |
TIMELESS interacting protein |
13649 |
0.11 |
| chr8_145002840_145002991 | 0.36 |
PLEC |
plectin |
1736 |
0.23 |
| chr8_37731176_37731327 | 0.36 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
4384 |
0.17 |
| chr12_32120805_32121101 | 0.36 |
KIAA1551 |
KIAA1551 |
5530 |
0.26 |
| chr4_124666055_124666206 | 0.36 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
345007 |
0.01 |
| chr6_147643085_147643755 | 0.36 |
STXBP5 |
syntaxin binding protein 5 (tomosyn) |
4889 |
0.34 |
| chr11_70163755_70163906 | 0.36 |
CTA-797E19.1 |
|
2204 |
0.21 |
| chr12_27391555_27391706 | 0.36 |
STK38L |
serine/threonine kinase 38 like |
5271 |
0.29 |
| chr10_26726645_26726796 | 0.36 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
412 |
0.9 |
| chr2_176868332_176868483 | 0.36 |
KIAA1715 |
KIAA1715 |
840 |
0.61 |
| chr2_7934753_7934904 | 0.35 |
ENSG00000221255 |
. |
217856 |
0.02 |
| chr10_4096081_4096232 | 0.35 |
KLF6 |
Kruppel-like factor 6 |
268683 |
0.02 |
| chr1_90223075_90223276 | 0.35 |
ENSG00000239176 |
. |
10702 |
0.21 |
| chr5_77805493_77806187 | 0.35 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
39134 |
0.21 |
| chr5_163343244_163343412 | 0.35 |
ENSG00000251998 |
. |
120157 |
0.07 |
| chr6_54063055_54063206 | 0.35 |
MLIP |
muscular LMNA-interacting protein |
86822 |
0.09 |
| chr2_238167076_238167227 | 0.35 |
AC112715.2 |
Uncharacterized protein |
1417 |
0.54 |
| chr11_104914891_104915295 | 0.35 |
CARD16 |
caspase recruitment domain family, member 16 |
941 |
0.54 |
| chr18_43470611_43470762 | 0.35 |
EPG5 |
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
22971 |
0.2 |
| chr5_133861629_133862874 | 0.35 |
JADE2 |
jade family PHD finger 2 |
76 |
0.97 |
| chr6_41996182_41996339 | 0.35 |
ENSG00000206875 |
. |
10008 |
0.15 |
| chr2_40621333_40621484 | 0.34 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
36012 |
0.23 |
| chrY_1550810_1551045 | 0.34 |
NA |
NA |
> 106 |
NA |
| chr19_54885564_54885768 | 0.34 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
3501 |
0.14 |
| chr5_88057335_88057486 | 0.34 |
MEF2C |
myocyte enhancer factor 2C |
62195 |
0.12 |
| chr3_37351986_37352375 | 0.34 |
GOLGA4 |
golgin A4 |
28506 |
0.15 |
| chr12_65032414_65032565 | 0.34 |
RP11-338E21.2 |
|
10365 |
0.12 |
| chr16_72965340_72965833 | 0.34 |
ENSG00000221799 |
. |
53170 |
0.13 |
| chr20_20760867_20761018 | 0.34 |
ENSG00000264361 |
. |
41568 |
0.19 |
| chr4_55575057_55575208 | 0.34 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
51047 |
0.18 |
| chr3_45729758_45730513 | 0.34 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
239 |
0.78 |
| chr20_5717977_5718128 | 0.34 |
C20orf196 |
chromosome 20 open reading frame 196 |
12987 |
0.26 |
| chr13_31312703_31312943 | 0.34 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
3178 |
0.36 |
| chrX_154027559_154027756 | 0.34 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
6015 |
0.13 |
| chr15_64984139_64984290 | 0.34 |
AC100830.4 |
|
3026 |
0.15 |
| chr8_97779057_97779208 | 0.34 |
CPQ |
carboxypeptidase Q |
5664 |
0.34 |
| chr19_54827917_54828068 | 0.34 |
LILRA5 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
3583 |
0.12 |
| chr4_139879629_139879991 | 0.34 |
RP11-371F15.3 |
|
20675 |
0.19 |
| chr17_65373728_65374565 | 0.33 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
125 |
0.95 |
| chr15_63539472_63539623 | 0.33 |
APH1B |
APH1B gamma secretase subunit |
28670 |
0.18 |
| chr10_134365004_134365155 | 0.33 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
13436 |
0.21 |
| chr5_75764900_75765051 | 0.33 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
64726 |
0.13 |
| chr8_24151584_24151953 | 0.33 |
ADAM28 |
ADAM metallopeptidase domain 28 |
144 |
0.97 |
| chr3_172230166_172230317 | 0.33 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
11024 |
0.25 |
| chr17_38443685_38443884 | 0.33 |
CDC6 |
cell division cycle 6 |
101 |
0.95 |
| chrX_9431281_9431551 | 0.33 |
TBL1X |
transducin (beta)-like 1X-linked |
45 |
0.99 |
| chr1_174934321_174935265 | 0.33 |
RABGAP1L |
RAB GTPase activating protein 1-like |
888 |
0.52 |
| chrX_39631447_39631598 | 0.33 |
ENSG00000264618 |
. |
14555 |
0.27 |
| chr14_64228721_64228872 | 0.32 |
ENSG00000252749 |
. |
25634 |
0.16 |
| chr18_60128151_60128302 | 0.32 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
62014 |
0.12 |
| chr9_101881133_101881552 | 0.32 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
8715 |
0.21 |
| chr1_26601594_26601745 | 0.32 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
3998 |
0.14 |
| chr8_139204133_139204284 | 0.32 |
FAM135B |
family with sequence similarity 135, member B |
233550 |
0.02 |
| chr6_13295173_13295324 | 0.32 |
RP1-257A7.4 |
|
570 |
0.76 |
| chr19_823853_824004 | 0.32 |
LPPR3 |
hsa-mir-3187 |
1961 |
0.15 |
| chr9_107826999_107827201 | 0.32 |
ENSG00000201583 |
. |
31937 |
0.22 |
| chr3_182978087_182978238 | 0.32 |
ENSG00000202502 |
. |
1958 |
0.34 |
| chr14_92652024_92652175 | 0.32 |
CPSF2 |
cleavage and polyadenylation specific factor 2, 100kDa |
30576 |
0.17 |
| chr19_51634092_51634243 | 0.32 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
3880 |
0.11 |
| chr12_1414903_1415054 | 0.32 |
RP5-951N9.2 |
|
80021 |
0.09 |
| chr10_81081747_81081898 | 0.31 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
15847 |
0.2 |
| chr6_43757843_43757994 | 0.31 |
VEGFA |
vascular endothelial growth factor A |
15828 |
0.18 |
| chr6_10837683_10838466 | 0.31 |
MAK |
male germ cell-associated kinase |
654 |
0.68 |
| chr1_65399507_65400011 | 0.31 |
JAK1 |
Janus kinase 1 |
32428 |
0.2 |
| chrX_40008490_40008641 | 0.31 |
BCOR |
BCL6 corepressor |
2683 |
0.42 |
| chr3_4793867_4794159 | 0.31 |
ENSG00000239126 |
. |
126373 |
0.05 |
| chr18_60920514_60920680 | 0.30 |
ENSG00000238988 |
. |
58699 |
0.1 |
| chr7_76829078_76829397 | 0.30 |
FGL2 |
fibrinogen-like 2 |
94 |
0.98 |
| chrX_153275945_153276096 | 0.30 |
IRAK1 |
interleukin-1 receptor-associated kinase 1 |
3677 |
0.13 |
| chr4_84188698_84188849 | 0.30 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
17145 |
0.23 |
| chr15_70755434_70755585 | 0.30 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
239111 |
0.02 |
| chr2_11971800_11971951 | 0.30 |
ENSG00000265172 |
. |
5237 |
0.22 |
| chr6_88609819_88609970 | 0.30 |
ENSG00000211983 |
. |
52544 |
0.13 |
| chrX_1600809_1601304 | 0.30 |
ASMTL |
acetylserotonin O-methyltransferase-like |
28401 |
0.16 |
| chr17_76748108_76748259 | 0.30 |
CYTH1 |
cytohesin 1 |
15211 |
0.18 |
| chr3_151960824_151960975 | 0.30 |
MBNL1 |
muscleblind-like splicing regulator 1 |
24930 |
0.2 |
| chr3_171853122_171853273 | 0.30 |
FNDC3B |
fibronectin type III domain containing 3B |
8433 |
0.29 |
| chr14_24743427_24743578 | 0.30 |
RABGGTA |
Rab geranylgeranyltransferase, alpha subunit |
2557 |
0.1 |
| chr5_149060859_149061010 | 0.30 |
ENSG00000221043 |
. |
14588 |
0.16 |
| chr10_28794428_28794579 | 0.30 |
WAC-AS1 |
WAC antisense RNA 1 (head to head) |
26780 |
0.17 |
| chrX_49853365_49853516 | 0.29 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
19284 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.2 | 0.7 | GO:0052033 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.2 | 0.6 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 | 0.5 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.2 | 0.5 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.4 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.4 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.4 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.1 | 0.2 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.2 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.1 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.1 | 0.2 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.1 | 0.3 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.2 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.1 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0052509 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.3 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.2 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0002246 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.5 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.0 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0052173 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.3 | GO:0018200 | peptidyl-glutamic acid modification(GO:0018200) |
| 0.0 | 0.1 | GO:0047496 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.0 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.0 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.0 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.1 | GO:0071027 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.0 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.6 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.1 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.0 | GO:0045630 | positive regulation of type 2 immune response(GO:0002830) positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.0 | GO:0010829 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.3 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.1 | 0.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.3 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.8 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) RNA polymerase II transcription cofactor activity(GO:0001104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.1 | 1.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 2.0 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |