Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
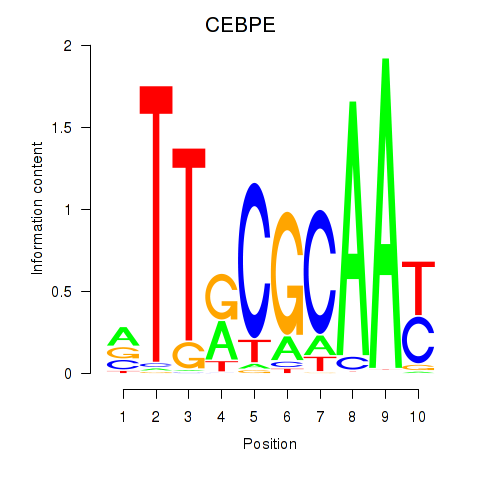
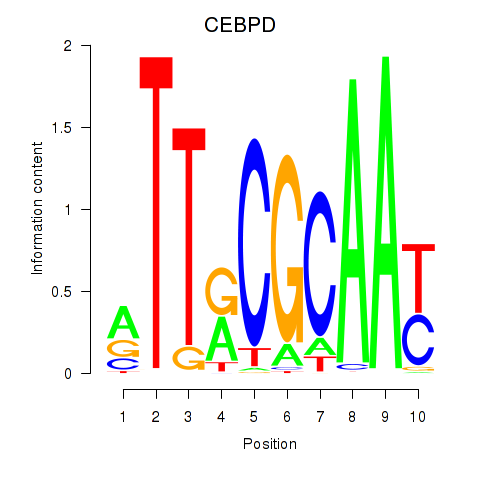
Results for CEBPE_CEBPD
Z-value: 2.16
Transcription factors associated with CEBPE_CEBPD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPE
|
ENSG00000092067.5 | CCAAT enhancer binding protein epsilon |
|
CEBPD
|
ENSG00000221869.4 | CCAAT enhancer binding protein delta |
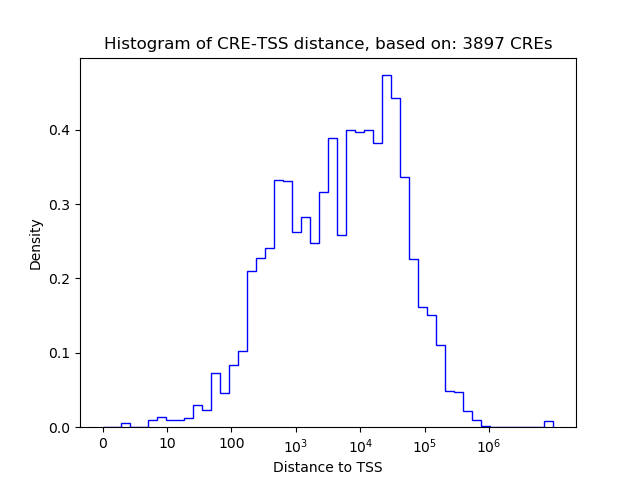
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_48647847_48648063 | CEBPD | 3693 | 0.222323 | 0.90 | 1.0e-03 | Click! |
| chr8_48648492_48649280 | CEBPD | 2762 | 0.255170 | 0.72 | 2.8e-02 | Click! |
| chr8_48648084_48648235 | CEBPD | 3489 | 0.227586 | 0.60 | 8.7e-02 | Click! |
| chr8_48651279_48651784 | CEBPD | 117 | 0.964335 | 0.56 | 1.1e-01 | Click! |
| chr8_48650852_48651279 | CEBPD | 583 | 0.743280 | 0.45 | 2.2e-01 | Click! |
| chr14_23583329_23583480 | CEBPE | 5421 | 0.108683 | 0.85 | 3.6e-03 | Click! |
| chr14_23587780_23587931 | CEBPE | 970 | 0.379591 | 0.81 | 8.8e-03 | Click! |
| chr14_23588022_23588173 | CEBPE | 728 | 0.496732 | 0.79 | 1.1e-02 | Click! |
| chr14_23589888_23590039 | CEBPE | 1138 | 0.323901 | 0.78 | 1.4e-02 | Click! |
| chr14_23589031_23589182 | CEBPE | 281 | 0.830672 | 0.73 | 2.4e-02 | Click! |
Activity of the CEBPE_CEBPD motif across conditions
Conditions sorted by the z-value of the CEBPE_CEBPD motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
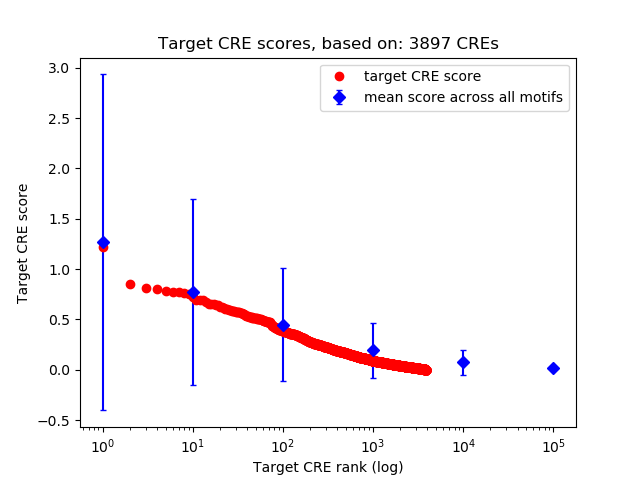
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_28619412_28619563 | 1.22 |
RP11-373D23.2 |
|
195 |
0.87 |
| chr17_78753891_78754042 | 0.86 |
RP11-28G8.1 |
|
25466 |
0.21 |
| chr11_59950051_59950297 | 0.81 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
325 |
0.9 |
| chr3_187607411_187607594 | 0.80 |
BCL6 |
B-cell CLL/lymphoma 6 |
143987 |
0.04 |
| chr11_32852041_32852192 | 0.78 |
PRRG4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
627 |
0.77 |
| chr14_93540505_93540656 | 0.78 |
ITPK1-AS1 |
ITPK1 antisense RNA 1 |
6783 |
0.21 |
| chr6_144357186_144358268 | 0.77 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
28008 |
0.21 |
| chr2_145444399_145444550 | 0.77 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
165853 |
0.04 |
| chr1_186649638_186650434 | 0.75 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
477 |
0.89 |
| chr20_54987227_54988129 | 0.73 |
CASS4 |
Cas scaffolding protein family member 4 |
361 |
0.82 |
| chr10_51575835_51576189 | 0.70 |
NCOA4 |
nuclear receptor coactivator 4 |
273 |
0.91 |
| chr11_1320381_1320746 | 0.70 |
TOLLIP |
toll interacting protein |
3457 |
0.18 |
| chr7_77168081_77168577 | 0.69 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
943 |
0.69 |
| chr2_219093418_219093644 | 0.67 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
11598 |
0.11 |
| chr2_136926324_136926475 | 0.65 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
50664 |
0.17 |
| chr8_56792958_56793527 | 0.65 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
848 |
0.55 |
| chrX_24498632_24498874 | 0.65 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
15180 |
0.2 |
| chr4_128553582_128553997 | 0.64 |
INTU |
inturned planar cell polarity protein |
298 |
0.94 |
| chr10_77531854_77532472 | 0.64 |
C10orf11 |
chromosome 10 open reading frame 11 |
10356 |
0.22 |
| chr2_152784984_152785135 | 0.63 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
43443 |
0.19 |
| chr5_36686005_36686156 | 0.62 |
CTD-2353F22.1 |
|
39195 |
0.2 |
| chr8_92044308_92044459 | 0.61 |
TMEM55A |
transmembrane protein 55A |
8680 |
0.13 |
| chr3_30673679_30673830 | 0.61 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
25661 |
0.24 |
| chr18_53255518_53256419 | 0.60 |
TCF4 |
transcription factor 4 |
108 |
0.98 |
| chr2_226983216_226983814 | 0.59 |
ENSG00000263363 |
. |
539994 |
0.0 |
| chr6_137180535_137180686 | 0.59 |
ENSG00000201807 |
. |
3641 |
0.24 |
| chr15_77861873_77862206 | 0.59 |
LINGO1 |
leucine rich repeat and Ig domain containing 1 |
62793 |
0.11 |
| chr9_139439215_139440028 | 0.59 |
NOTCH1 |
notch 1 |
693 |
0.35 |
| chr3_52236882_52237267 | 0.58 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
3574 |
0.14 |
| chr14_99541797_99541948 | 0.58 |
AL162151.4 |
|
82881 |
0.1 |
| chr15_94716341_94716492 | 0.57 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
58351 |
0.16 |
| chr15_42803128_42803279 | 0.57 |
SNAP23 |
synaptosomal-associated protein, 23kDa |
842 |
0.51 |
| chr17_57803272_57803423 | 0.57 |
VMP1 |
vacuole membrane protein 1 |
3729 |
0.21 |
| chr7_36763519_36763817 | 0.57 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
386 |
0.9 |
| chr17_25867508_25867659 | 0.56 |
KSR1 |
kinase suppressor of ras 1 |
42364 |
0.14 |
| chr4_70579445_70579596 | 0.56 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
46796 |
0.14 |
| chr14_21387247_21387398 | 0.55 |
RNASE3 |
ribonuclease, RNase A family, 3 |
27764 |
0.1 |
| chr1_235012225_235012376 | 0.55 |
ENSG00000239690 |
. |
27633 |
0.2 |
| chr4_154582407_154582558 | 0.54 |
RP11-153M7.3 |
|
16929 |
0.19 |
| chr2_174889838_174890106 | 0.54 |
SP3 |
Sp3 transcription factor |
59542 |
0.15 |
| chr9_95491243_95491394 | 0.53 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
35776 |
0.15 |
| chr4_74570333_74570484 | 0.53 |
IL8 |
interleukin 8 |
35815 |
0.18 |
| chr2_113597126_113597277 | 0.52 |
IL1B |
interleukin 1, beta |
2721 |
0.23 |
| chr18_74277400_74278128 | 0.52 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
36890 |
0.17 |
| chr7_29603979_29604650 | 0.52 |
PRR15 |
proline rich 15 |
752 |
0.51 |
| chr16_19524952_19525190 | 0.51 |
GDE1 |
glycerophosphodiester phosphodiesterase 1 |
3419 |
0.15 |
| chr17_75425897_75426048 | 0.51 |
SEPT9 |
septin 9 |
2934 |
0.21 |
| chr12_122269898_122270049 | 0.51 |
RP11-7M8.2 |
|
24192 |
0.12 |
| chr13_77319930_77320170 | 0.51 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
140475 |
0.05 |
| chr8_134258257_134258408 | 0.51 |
WISP1-OT1 |
WISP1 overlapping transcript 1 (non-protein coding) |
17033 |
0.22 |
| chr5_147845522_147845673 | 0.51 |
CTD-2283N19.1 |
|
35227 |
0.17 |
| chr2_169926772_169926923 | 0.51 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
761 |
0.69 |
| chr1_40400191_40400342 | 0.51 |
ENSG00000207356 |
. |
10404 |
0.13 |
| chr1_16212355_16212620 | 0.51 |
SPEN |
spen family transcriptional repressor |
9416 |
0.15 |
| chr12_76937269_76937420 | 0.50 |
OSBPL8 |
oxysterol binding protein-like 8 |
15940 |
0.29 |
| chr7_50859466_50860386 | 0.50 |
GRB10 |
growth factor receptor-bound protein 10 |
740 |
0.77 |
| chr11_118097499_118097650 | 0.50 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
1765 |
0.28 |
| chr1_243430959_243431113 | 0.50 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
11664 |
0.19 |
| chr17_592967_593118 | 0.49 |
VPS53 |
vacuolar protein sorting 53 homolog (S. cerevisiae) |
24907 |
0.15 |
| chr12_27242472_27242623 | 0.49 |
C12orf71 |
chromosome 12 open reading frame 71 |
7100 |
0.23 |
| chr17_61043904_61045029 | 0.49 |
ENSG00000207552 |
. |
22890 |
0.21 |
| chr14_73928931_73929214 | 0.49 |
ENSG00000251393 |
. |
57 |
0.97 |
| chr15_63780195_63780346 | 0.49 |
USP3 |
ubiquitin specific peptidase 3 |
16523 |
0.23 |
| chr1_167570877_167571161 | 0.49 |
RCSD1 |
RCSD domain containing 1 |
28311 |
0.15 |
| chrX_9320798_9321168 | 0.48 |
TBL1X |
transducin (beta)-like 1X-linked |
110352 |
0.07 |
| chr19_7799235_7799386 | 0.48 |
CLEC4G |
C-type lectin domain family 4, member G |
518 |
0.64 |
| chr12_11803481_11804350 | 0.48 |
ETV6 |
ets variant 6 |
1127 |
0.62 |
| chr16_28186650_28186801 | 0.47 |
ENSG00000200652 |
. |
3694 |
0.2 |
| chr15_64456219_64456370 | 0.47 |
PPIB |
peptidylprolyl isomerase B (cyclophilin B) |
890 |
0.54 |
| chr8_9409853_9410128 | 0.47 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
3434 |
0.27 |
| chr11_63639674_63640171 | 0.47 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
3174 |
0.18 |
| chr9_123833984_123834761 | 0.47 |
CNTRL |
centriolin |
2769 |
0.29 |
| chr14_75920844_75920995 | 0.46 |
JDP2 |
Jun dimerization protein 2 |
22082 |
0.17 |
| chr1_41326478_41326629 | 0.46 |
CITED4 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
1465 |
0.35 |
| chr22_35805907_35806058 | 0.45 |
MCM5 |
minichromosome maintenance complex component 5 |
9558 |
0.2 |
| chr9_134268612_134269131 | 0.44 |
PRRC2B |
proline-rich coiled-coil 2B |
609 |
0.77 |
| chr10_4173141_4173857 | 0.43 |
KLF6 |
Kruppel-like factor 6 |
346026 |
0.01 |
| chrX_15518847_15518998 | 0.43 |
BMX |
BMX non-receptor tyrosine kinase |
30 |
0.98 |
| chr13_103333518_103333882 | 0.43 |
METTL21C |
methyltransferase like 21C |
13154 |
0.16 |
| chr10_112033444_112033726 | 0.42 |
SMNDC1 |
survival motor neuron domain containing 1 |
30870 |
0.18 |
| chr17_36715792_36715943 | 0.42 |
SRCIN1 |
SRC kinase signaling inhibitor 1 |
3793 |
0.19 |
| chr12_94241991_94242142 | 0.42 |
RP11-887P2.6 |
|
12123 |
0.21 |
| chr19_7710267_7710418 | 0.42 |
STXBP2 |
syntaxin binding protein 2 |
503 |
0.58 |
| chr3_171779467_171780048 | 0.42 |
FNDC3B |
fibronectin type III domain containing 3B |
15054 |
0.27 |
| chr5_147566124_147566390 | 0.42 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
16100 |
0.18 |
| chr11_10329247_10329398 | 0.42 |
ADM |
adrenomedullin |
2398 |
0.23 |
| chr3_66514964_66515132 | 0.41 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
36308 |
0.22 |
| chr13_49141961_49142410 | 0.40 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
34816 |
0.21 |
| chr21_35917446_35917660 | 0.40 |
RCAN1 |
regulator of calcineurin 1 |
14924 |
0.2 |
| chr15_78305137_78305288 | 0.40 |
TBC1D2B |
TBC1 domain family, member 2B |
11186 |
0.13 |
| chr16_28610570_28610819 | 0.40 |
SULT1A2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
2264 |
0.2 |
| chr16_72088411_72088562 | 0.40 |
HP |
haptoglobin |
5 |
0.51 |
| chr19_31820042_31820193 | 0.40 |
AC007796.1 |
|
19670 |
0.23 |
| chr21_36880734_36880885 | 0.40 |
ENSG00000211590 |
. |
212204 |
0.02 |
| chr10_65015927_65016153 | 0.39 |
JMJD1C |
jumonji domain containing 1C |
12786 |
0.27 |
| chr17_80229846_80229997 | 0.39 |
CSNK1D |
casein kinase 1, delta |
1383 |
0.28 |
| chr17_7462231_7462382 | 0.39 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
202 |
0.78 |
| chr1_12046097_12046354 | 0.39 |
MFN2 |
mitofusin 2 |
4176 |
0.15 |
| chr16_642735_643008 | 0.39 |
RAB40C |
RAB40C, member RAS oncogene family |
2539 |
0.11 |
| chr1_153330986_153331137 | 0.39 |
S100A9 |
S100 calcium binding protein A9 |
731 |
0.51 |
| chr17_76588567_76588821 | 0.38 |
DNAH17 |
dynein, axonemal, heavy chain 17 |
15218 |
0.19 |
| chr1_200558260_200558411 | 0.38 |
KIF14 |
kinesin family member 14 |
31527 |
0.2 |
| chr2_185463650_185463898 | 0.38 |
ZNF804A |
zinc finger protein 804A |
681 |
0.83 |
| chr1_36521425_36521576 | 0.38 |
TEKT2 |
tektin 2 (testicular) |
28176 |
0.13 |
| chr20_17663271_17663734 | 0.38 |
RRBP1 |
ribosome binding protein 1 |
562 |
0.78 |
| chr3_98451551_98452443 | 0.38 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
425 |
0.53 |
| chr9_140502849_140503000 | 0.38 |
ARRDC1 |
arrestin domain containing 1 |
2769 |
0.16 |
| chr20_43610553_43610704 | 0.38 |
STK4 |
serine/threonine kinase 4 |
15461 |
0.14 |
| chr2_124667522_124667673 | 0.38 |
ENSG00000223118 |
. |
40450 |
0.2 |
| chr10_111827720_111827871 | 0.37 |
ADD3 |
adducin 3 (gamma) |
60073 |
0.12 |
| chr15_42349751_42349902 | 0.37 |
PLA2G4E |
phospholipase A2, group IVE |
6438 |
0.17 |
| chr1_38317055_38317206 | 0.37 |
MTF1 |
metal-regulatory transcription factor 1 |
8162 |
0.1 |
| chr1_180982364_180982515 | 0.37 |
STX6 |
syntaxin 6 |
9566 |
0.16 |
| chr3_72944167_72944405 | 0.37 |
GXYLT2 |
glucoside xylosyltransferase 2 |
7062 |
0.24 |
| chr10_47670479_47670739 | 0.37 |
ANTXRL |
anthrax toxin receptor-like |
12360 |
0.2 |
| chr17_73089180_73089331 | 0.37 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
127 |
0.91 |
| chr21_44035208_44035359 | 0.36 |
AP001626.2 |
|
36071 |
0.13 |
| chr12_49628889_49629058 | 0.36 |
TUBA1C |
tubulin, alpha 1c |
7264 |
0.13 |
| chr2_64500730_64500881 | 0.36 |
AC074289.1 |
|
45203 |
0.16 |
| chr8_22961570_22961743 | 0.36 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
1222 |
0.37 |
| chr5_40678717_40679405 | 0.36 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
539 |
0.8 |
| chr17_38269362_38269513 | 0.36 |
MSL1 |
male-specific lethal 1 homolog (Drosophila) |
9114 |
0.11 |
| chr5_49944181_49944332 | 0.36 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
17477 |
0.3 |
| chr2_123738319_123738470 | 0.36 |
ENSG00000221689 |
. |
701397 |
0.0 |
| chr17_40463640_40463791 | 0.36 |
STAT5A |
signal transducer and activator of transcription 5A |
5527 |
0.14 |
| chr3_146260050_146260212 | 0.36 |
PLSCR1 |
phospholipid scramblase 1 |
2177 |
0.31 |
| chr15_80271248_80271486 | 0.36 |
BCL2A1 |
BCL2-related protein A1 |
7579 |
0.2 |
| chr7_79408525_79408693 | 0.36 |
ENSG00000200786 |
. |
125184 |
0.06 |
| chr1_26662362_26662902 | 0.35 |
AIM1L |
absent in melanoma 1-like |
3205 |
0.15 |
| chr1_87796860_87797240 | 0.35 |
LMO4 |
LIM domain only 4 |
301 |
0.95 |
| chr20_58513678_58513898 | 0.35 |
PPP1R3D |
protein phosphatase 1, regulatory subunit 3D |
1564 |
0.28 |
| chr18_74872659_74872810 | 0.35 |
MBP |
myelin basic protein |
27934 |
0.24 |
| chr18_9779691_9779842 | 0.35 |
ENSG00000242651 |
. |
50858 |
0.12 |
| chr12_89746565_89746947 | 0.35 |
DUSP6 |
dual specificity phosphatase 6 |
292 |
0.94 |
| chr2_219671245_219671396 | 0.35 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
683 |
0.61 |
| chr10_125851443_125851830 | 0.35 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
334 |
0.93 |
| chr3_58320901_58321052 | 0.35 |
PXK |
PX domain containing serine/threonine kinase |
1943 |
0.34 |
| chr1_27951756_27951907 | 0.35 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
815 |
0.54 |
| chr7_115671621_115671772 | 0.35 |
TFEC |
transcription factor EC |
829 |
0.76 |
| chr2_29175549_29175700 | 0.34 |
FAM179A |
family with sequence similarity 179, member A |
3853 |
0.14 |
| chr6_134343019_134343170 | 0.34 |
SLC2A12 |
solute carrier family 2 (facilitated glucose transporter), member 12 |
30680 |
0.19 |
| chr4_113066984_113067681 | 0.34 |
C4orf32 |
chromosome 4 open reading frame 32 |
779 |
0.73 |
| chr17_25609205_25609356 | 0.34 |
ENSG00000263583 |
. |
11742 |
0.23 |
| chr4_86396251_86396726 | 0.34 |
ARHGAP24 |
Rho GTPase activating protein 24 |
97 |
0.99 |
| chr6_146867809_146867960 | 0.34 |
RAB32 |
RAB32, member RAS oncogene family |
3055 |
0.34 |
| chr9_21023989_21024197 | 0.34 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
7515 |
0.22 |
| chr4_157115087_157115238 | 0.34 |
ENSG00000221189 |
. |
102636 |
0.08 |
| chr13_48736574_48736800 | 0.34 |
MED4 |
mediator complex subunit 4 |
67420 |
0.1 |
| chr10_3710614_3710914 | 0.34 |
RP11-184A2.3 |
|
82495 |
0.1 |
| chr16_4103676_4103973 | 0.33 |
RP11-462G12.4 |
|
21811 |
0.19 |
| chr17_55980824_55981073 | 0.33 |
CUEDC1 |
CUE domain containing 1 |
198 |
0.94 |
| chr19_52239080_52239231 | 0.33 |
HAS1 |
hyaluronan synthase 1 |
11908 |
0.1 |
| chr8_20057882_20058033 | 0.33 |
ATP6V1B2 |
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
3007 |
0.25 |
| chr8_17193981_17194132 | 0.32 |
MTMR7 |
myotubularin related protein 7 |
24874 |
0.21 |
| chr8_129262697_129262848 | 0.32 |
ENSG00000201782 |
. |
30022 |
0.23 |
| chr17_76912321_76912483 | 0.32 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
6081 |
0.15 |
| chr3_120277883_120278034 | 0.32 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
37198 |
0.19 |
| chr16_11678014_11678655 | 0.32 |
LITAF |
lipopolysaccharide-induced TNF factor |
1895 |
0.35 |
| chr3_43221570_43221860 | 0.32 |
ENSG00000222331 |
. |
30648 |
0.18 |
| chr13_110357339_110357490 | 0.32 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
23215 |
0.26 |
| chr12_81102183_81102935 | 0.32 |
MYF6 |
myogenic factor 6 (herculin) |
1282 |
0.49 |
| chr4_106035170_106035321 | 0.32 |
ENSG00000252136 |
. |
8101 |
0.23 |
| chr15_57595155_57595306 | 0.32 |
TCF12 |
transcription factor 12 |
20577 |
0.2 |
| chr13_48796681_48797014 | 0.32 |
ITM2B |
integral membrane protein 2B |
10447 |
0.26 |
| chr2_163199250_163199401 | 0.32 |
GCA |
grancalcin, EF-hand calcium binding protein |
1273 |
0.52 |
| chr9_126629276_126629488 | 0.32 |
DENND1A |
DENN/MADD domain containing 1A |
63004 |
0.12 |
| chr12_68758221_68758372 | 0.32 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
32135 |
0.22 |
| chr15_65277376_65277561 | 0.31 |
SPG21 |
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
2346 |
0.25 |
| chr17_77789018_77789210 | 0.31 |
ENSG00000238331 |
. |
4636 |
0.15 |
| chr5_43040634_43042182 | 0.31 |
AC025171.1 |
|
269 |
0.87 |
| chr2_74765980_74766272 | 0.31 |
HTRA2 |
HtrA serine peptidase 2 |
8953 |
0.07 |
| chr8_12615101_12615641 | 0.31 |
LONRF1 |
LON peptidase N-terminal domain and ring finger 1 |
2372 |
0.31 |
| chr11_104904746_104905919 | 0.31 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
508 |
0.77 |
| chr12_123376332_123377156 | 0.31 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
2038 |
0.29 |
| chr8_26370640_26370791 | 0.30 |
PNMA2 |
paraneoplastic Ma antigen 2 |
893 |
0.46 |
| chr6_53661674_53661825 | 0.30 |
LRRC1 |
leucine rich repeat containing 1 |
1861 |
0.34 |
| chr10_99223651_99223802 | 0.30 |
MMS19 |
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
3556 |
0.12 |
| chr9_94184850_94185394 | 0.30 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
1022 |
0.67 |
| chr3_172240144_172241280 | 0.30 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
553 |
0.83 |
| chr3_171788685_171788836 | 0.30 |
FNDC3B |
fibronectin type III domain containing 3B |
24057 |
0.25 |
| chr14_21738093_21738425 | 0.29 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
606 |
0.71 |
| chr11_48075840_48075991 | 0.29 |
AC103828.1 |
|
38508 |
0.15 |
| chr3_59119295_59119446 | 0.29 |
C3orf67 |
chromosome 3 open reading frame 67 |
83560 |
0.11 |
| chr15_93460792_93461223 | 0.29 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
13304 |
0.19 |
| chr1_109913452_109914259 | 0.29 |
SORT1 |
sortilin 1 |
22124 |
0.14 |
| chr22_48554335_48554486 | 0.29 |
ENSG00000266508 |
. |
115766 |
0.07 |
| chr2_143904464_143904615 | 0.29 |
ARHGAP15 |
Rho GTPase activating protein 15 |
17656 |
0.23 |
| chr8_126503508_126503659 | 0.29 |
ENSG00000266452 |
. |
46776 |
0.17 |
| chr13_33001190_33001359 | 0.29 |
N4BP2L1 |
NEDD4 binding protein 2-like 1 |
877 |
0.62 |
| chr7_106807796_106808590 | 0.29 |
HBP1 |
HMG-box transcription factor 1 |
1213 |
0.5 |
| chr2_85815414_85815565 | 0.29 |
VAMP5 |
vesicle-associated membrane protein 5 |
3958 |
0.1 |
| chr17_26385024_26385175 | 0.28 |
ENSG00000252283 |
. |
3894 |
0.19 |
| chr6_112194486_112194668 | 0.28 |
FYN |
FYN oncogene related to SRC, FGR, YES |
50 |
0.99 |
| chr9_128521797_128522422 | 0.28 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
11631 |
0.27 |
| chr10_17259722_17259873 | 0.28 |
VIM-AS1 |
VIM antisense RNA 1 |
9100 |
0.16 |
| chr10_27736608_27736759 | 0.28 |
PTCHD3 |
patched domain containing 3 |
33386 |
0.18 |
| chr11_46378662_46378813 | 0.28 |
DGKZ |
diacylglycerol kinase, zeta |
4408 |
0.16 |
| chr8_30581756_30582064 | 0.28 |
GSR |
glutathione reductase |
2617 |
0.31 |
| chr2_201987732_201987883 | 0.28 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
607 |
0.65 |
| chr3_101568772_101569567 | 0.28 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
62 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.6 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.5 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.1 | 0.4 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.1 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.1 | 0.4 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.5 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.1 | 0.2 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.3 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.2 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.1 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.3 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.2 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.2 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.0 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0052553 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.1 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.4 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0090195 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.2 | GO:0055093 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.9 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0043300 | regulation of leukocyte degranulation(GO:0043300) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.4 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0044409 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.0 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.7 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of humoral immune response(GO:0002921) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0090312 | regulation of protein deacetylation(GO:0090311) positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.3 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0036230 | granulocyte activation(GO:0036230) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.0 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:1903077 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0010042 | response to manganese ion(GO:0010042) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.2 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.0 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.0 | 0.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0006907 | pinocytosis(GO:0006907) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 1.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.0 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.2 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.6 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.3 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 1.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.6 | GO:0016684 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0051192 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |