Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
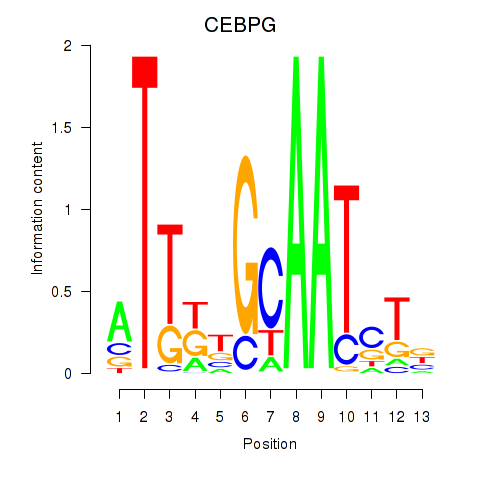
Results for CEBPG
Z-value: 1.03
Transcription factors associated with CEBPG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPG
|
ENSG00000153879.4 | CCAAT enhancer binding protein gamma |
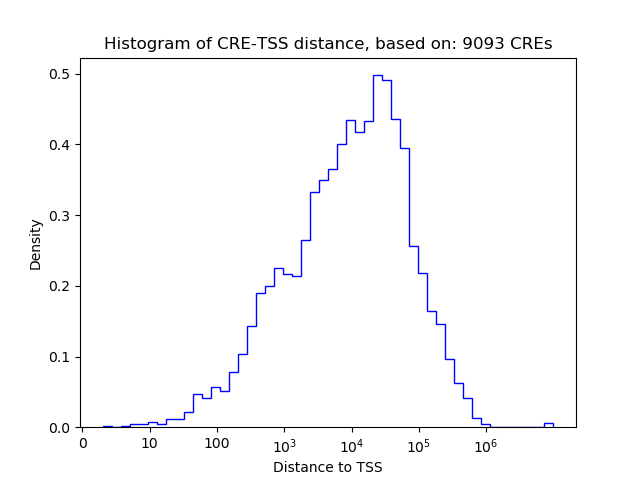
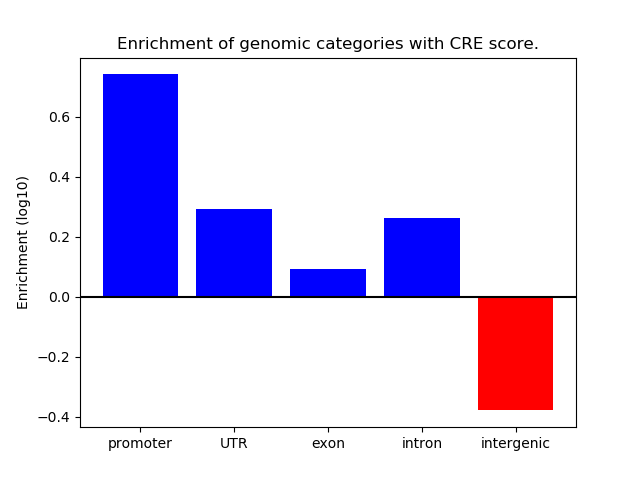
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_33866567_33866804 | CEBPG | 1467 | 0.426703 | 0.75 | 1.9e-02 | Click! |
| chr19_33865170_33865786 | CEBPG | 260 | 0.924510 | -0.71 | 3.1e-02 | Click! |
| chr19_33867678_33867829 | CEBPG | 2535 | 0.292984 | 0.71 | 3.1e-02 | Click! |
| chr19_33870618_33870769 | CEBPG | 5475 | 0.220899 | 0.60 | 8.7e-02 | Click! |
| chr19_33885155_33885306 | CEBPG | 20012 | 0.188837 | 0.51 | 1.6e-01 | Click! |
Activity of the CEBPG motif across conditions
Conditions sorted by the z-value of the CEBPG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
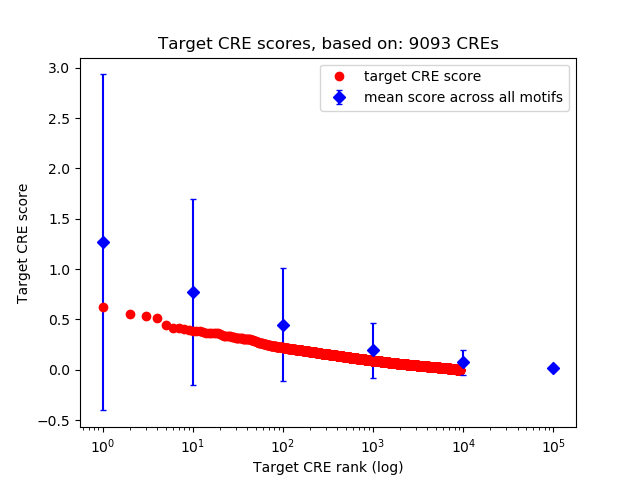
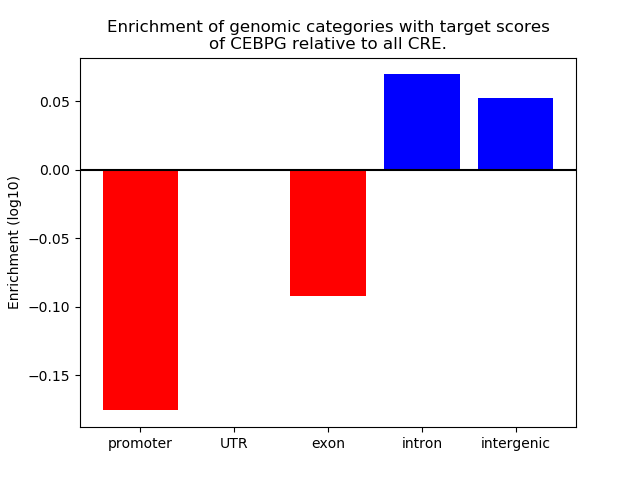
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_56792958_56793527 | 0.63 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
848 |
0.55 |
| chr11_78271810_78272407 | 0.56 |
NARS2 |
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
12467 |
0.18 |
| chr17_64464860_64465690 | 0.54 |
RP11-4F22.2 |
|
52303 |
0.15 |
| chr4_105982764_105983024 | 0.51 |
ENSG00000252136 |
. |
44250 |
0.17 |
| chr18_74179382_74179755 | 0.44 |
ZNF516 |
zinc finger protein 516 |
23167 |
0.16 |
| chr7_37771982_37772133 | 0.42 |
GPR141 |
G protein-coupled receptor 141 |
7939 |
0.27 |
| chr12_81102183_81102935 | 0.41 |
MYF6 |
myogenic factor 6 (herculin) |
1282 |
0.49 |
| chr2_201958459_201958610 | 0.40 |
ENSG00000252148 |
. |
13969 |
0.12 |
| chr4_157692811_157693135 | 0.39 |
RP11-154F14.2 |
|
69538 |
0.11 |
| chr2_238889404_238889624 | 0.39 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
7777 |
0.21 |
| chr19_49864989_49865626 | 0.38 |
DKKL1 |
dickkopf-like 1 |
267 |
0.51 |
| chr12_52427316_52427521 | 0.38 |
NR4A1 |
nuclear receptor subfamily 4, group A, member 1 |
3543 |
0.16 |
| chr13_27334023_27334664 | 0.38 |
GPR12 |
G protein-coupled receptor 12 |
84 |
0.98 |
| chr20_19983837_19984098 | 0.37 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
13793 |
0.21 |
| chr11_35949210_35949523 | 0.37 |
RP11-698N11.2 |
|
9077 |
0.19 |
| chr7_155446675_155446826 | 0.36 |
RBM33 |
RNA binding motif protein 33 |
9354 |
0.22 |
| chr5_94417062_94417734 | 0.36 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
48 |
0.99 |
| chr10_14606601_14606845 | 0.36 |
FAM107B |
family with sequence similarity 107, member B |
7306 |
0.26 |
| chr7_33089731_33090334 | 0.36 |
ENSG00000241420 |
. |
4330 |
0.18 |
| chr2_163199250_163199401 | 0.36 |
GCA |
grancalcin, EF-hand calcium binding protein |
1273 |
0.52 |
| chr15_99192776_99194432 | 0.34 |
IGF1R |
insulin-like growth factor 1 receptor |
1331 |
0.47 |
| chr3_11758192_11758601 | 0.34 |
VGLL4 |
vestigial like 4 (Drosophila) |
2672 |
0.38 |
| chr15_70052729_70052912 | 0.34 |
ENSG00000215958 |
. |
25023 |
0.21 |
| chr20_45980052_45980426 | 0.34 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
382 |
0.71 |
| chr11_63639674_63640171 | 0.33 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
3174 |
0.18 |
| chr2_226983216_226983814 | 0.33 |
ENSG00000263363 |
. |
539994 |
0.0 |
| chr5_58592476_58593460 | 0.32 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
21023 |
0.29 |
| chr2_230271788_230271939 | 0.32 |
PID1 |
phosphotyrosine interaction domain containing 1 |
135862 |
0.05 |
| chr5_153424892_153425043 | 0.32 |
MFAP3 |
microfibrillar-associated protein 3 |
6408 |
0.22 |
| chr15_83501820_83502050 | 0.32 |
HOMER2 |
homer homolog 2 (Drosophila) |
16544 |
0.13 |
| chr19_33790953_33791293 | 0.32 |
CTD-2540B15.11 |
|
283 |
0.79 |
| chr10_51575835_51576189 | 0.31 |
NCOA4 |
nuclear receptor coactivator 4 |
273 |
0.91 |
| chr11_59319331_59319576 | 0.31 |
ENSG00000252893 |
. |
6820 |
0.14 |
| chr1_36500237_36500507 | 0.31 |
RP4-665N4.8 |
|
21141 |
0.16 |
| chr5_53813164_53813315 | 0.31 |
SNX18 |
sorting nexin 18 |
350 |
0.92 |
| chr18_9673690_9673841 | 0.31 |
RAB31 |
RAB31, member RAS oncogene family |
34397 |
0.15 |
| chr9_93564472_93565060 | 0.31 |
SYK |
spleen tyrosine kinase |
557 |
0.87 |
| chr19_16259637_16259788 | 0.31 |
HSH2D |
hematopoietic SH2 domain containing |
5166 |
0.13 |
| chr7_12609670_12609821 | 0.31 |
SCIN |
scinderin |
567 |
0.7 |
| chr9_110495286_110496024 | 0.30 |
AL162389.1 |
Uncharacterized protein |
44764 |
0.15 |
| chr1_108479342_108479642 | 0.30 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
27573 |
0.19 |
| chr9_89952337_89952906 | 0.30 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr10_111827720_111827871 | 0.30 |
ADD3 |
adducin 3 (gamma) |
60073 |
0.12 |
| chr10_17241830_17242242 | 0.30 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
1543 |
0.37 |
| chr11_46181275_46181426 | 0.30 |
PHF21A |
PHD finger protein 21A |
38365 |
0.15 |
| chr2_63287244_63287598 | 0.30 |
OTX1 |
orthodenticle homeobox 1 |
9484 |
0.23 |
| chr1_12046097_12046354 | 0.30 |
MFN2 |
mitofusin 2 |
4176 |
0.15 |
| chr2_64500730_64500881 | 0.29 |
AC074289.1 |
|
45203 |
0.16 |
| chr10_3814711_3815005 | 0.29 |
RP11-184A2.2 |
|
4751 |
0.24 |
| chr10_5491792_5492244 | 0.28 |
NET1 |
neuroepithelial cell transforming 1 |
3444 |
0.22 |
| chr3_4026218_4026369 | 0.28 |
LRRN1 |
leucine rich repeat neuronal 1 |
185172 |
0.03 |
| chr1_94882933_94883167 | 0.27 |
ABCD3 |
ATP-binding cassette, sub-family D (ALD), member 3 |
883 |
0.72 |
| chr5_88178054_88178448 | 0.27 |
MEF2C |
myocyte enhancer factor 2C |
713 |
0.52 |
| chr1_221247887_221248144 | 0.27 |
HLX |
H2.0-like homeobox |
193431 |
0.03 |
| chr13_48796681_48797014 | 0.27 |
ITM2B |
integral membrane protein 2B |
10447 |
0.26 |
| chr22_39808489_39808802 | 0.27 |
TAB1 |
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
12856 |
0.15 |
| chr6_25003002_25003351 | 0.26 |
ENSG00000244618 |
. |
28334 |
0.16 |
| chr1_42230520_42230906 | 0.26 |
ENSG00000264896 |
. |
5901 |
0.3 |
| chr2_851043_851194 | 0.26 |
AC113607.3 |
|
65692 |
0.12 |
| chr9_110310779_110311295 | 0.26 |
KLF4 |
Kruppel-like factor 4 (gut) |
58274 |
0.13 |
| chr21_45509274_45509425 | 0.26 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
17822 |
0.16 |
| chrX_18686331_18686482 | 0.26 |
RS1 |
retinoschisin 1 |
3823 |
0.25 |
| chr2_176971683_176972255 | 0.26 |
HOXD11 |
homeobox D11 |
45 |
0.92 |
| chr10_26985869_26986716 | 0.26 |
PDSS1 |
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
296 |
0.92 |
| chr7_2851005_2851785 | 0.25 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
3497 |
0.32 |
| chr15_68497324_68497660 | 0.25 |
CALML4 |
calmodulin-like 4 |
334 |
0.87 |
| chr20_58547296_58547466 | 0.25 |
CDH26 |
cadherin 26 |
13899 |
0.17 |
| chr19_8750057_8750208 | 0.25 |
ACTL9 |
actin-like 9 |
59040 |
0.09 |
| chr17_68166405_68166607 | 0.25 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
830 |
0.45 |
| chr9_128521797_128522422 | 0.24 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
11631 |
0.27 |
| chr5_77784955_77785106 | 0.24 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
59944 |
0.15 |
| chr8_123256270_123256596 | 0.24 |
ENSG00000238901 |
. |
427097 |
0.01 |
| chr8_26288224_26288375 | 0.24 |
RP11-14I17.2 |
|
8190 |
0.19 |
| chr8_16689434_16690090 | 0.24 |
ENSG00000264092 |
. |
31243 |
0.24 |
| chr11_116856860_116857090 | 0.24 |
SIK3 |
SIK family kinase 3 |
29215 |
0.13 |
| chr5_16509247_16509418 | 0.24 |
FAM134B |
family with sequence similarity 134, member B |
225 |
0.95 |
| chr7_27290832_27290983 | 0.24 |
EVX1-AS |
EVX1 antisense RNA |
4059 |
0.14 |
| chr4_84161720_84161871 | 0.23 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
44123 |
0.16 |
| chr19_4374282_4374693 | 0.23 |
SH3GL1 |
SH3-domain GRB2-like 1 |
5802 |
0.08 |
| chr9_124261192_124261996 | 0.23 |
ENSG00000240299 |
. |
4797 |
0.23 |
| chr9_70850276_70850542 | 0.23 |
CBWD3 |
COBW domain containing 3 |
5988 |
0.18 |
| chr10_101605795_101606213 | 0.23 |
DNMBP |
dynamin binding protein |
52015 |
0.12 |
| chr3_138892567_138893067 | 0.23 |
MRPS22 |
mitochondrial ribosomal protein S22 |
63571 |
0.12 |
| chr5_173334437_173334588 | 0.23 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
14431 |
0.25 |
| chr16_21620226_21620509 | 0.23 |
METTL9 |
methyltransferase like 9 |
3054 |
0.17 |
| chr12_10874089_10875353 | 0.23 |
YBX3 |
Y box binding protein 3 |
1185 |
0.46 |
| chr19_38756015_38756259 | 0.23 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
159 |
0.9 |
| chr14_67709110_67709261 | 0.23 |
MPP5 |
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
841 |
0.64 |
| chr5_154145914_154146065 | 0.23 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
9255 |
0.18 |
| chr5_148207768_148208716 | 0.23 |
ADRB2 |
adrenoceptor beta 2, surface |
2086 |
0.43 |
| chr5_26247860_26248011 | 0.23 |
ENSG00000222560 |
. |
234937 |
0.02 |
| chr7_28705588_28705739 | 0.23 |
CREB5 |
cAMP responsive element binding protein 5 |
19935 |
0.29 |
| chr16_89600841_89601123 | 0.22 |
ENSG00000252579 |
. |
1020 |
0.38 |
| chr7_139561657_139561808 | 0.22 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
32622 |
0.21 |
| chr5_52775201_52775957 | 0.22 |
FST |
follistatin |
660 |
0.81 |
| chr14_51289214_51289365 | 0.22 |
NIN |
ninein (GSK3B interacting protein) |
515 |
0.52 |
| chr8_126290495_126290646 | 0.22 |
ENSG00000242170 |
. |
7724 |
0.27 |
| chr5_119542834_119542985 | 0.22 |
ENSG00000251975 |
. |
130439 |
0.06 |
| chr17_1458782_1458933 | 0.22 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
2491 |
0.19 |
| chr1_61668583_61669016 | 0.22 |
RP4-802A10.1 |
|
78394 |
0.11 |
| chr10_1090354_1090600 | 0.22 |
ENSG00000238924 |
. |
2182 |
0.2 |
| chr6_88407613_88407764 | 0.22 |
AKIRIN2 |
akirin 2 |
4239 |
0.23 |
| chr3_171780331_171780482 | 0.22 |
FNDC3B |
fibronectin type III domain containing 3B |
15703 |
0.27 |
| chr7_70201396_70202343 | 0.22 |
AUTS2 |
autism susceptibility candidate 2 |
7744 |
0.34 |
| chr4_172263864_172264142 | 0.22 |
ENSG00000251961 |
. |
451385 |
0.01 |
| chr4_91348946_91349097 | 0.22 |
ENSG00000252087 |
. |
57747 |
0.17 |
| chr16_53899292_53899443 | 0.22 |
FTO |
fat mass and obesity associated |
21462 |
0.24 |
| chr4_120291951_120292186 | 0.22 |
ENSG00000201186 |
. |
3351 |
0.26 |
| chr8_32165347_32165498 | 0.22 |
ENSG00000200246 |
. |
51410 |
0.15 |
| chr6_133034939_133035106 | 0.21 |
VNN1 |
vanin 1 |
166 |
0.94 |
| chr2_147579520_147579813 | 0.21 |
ENSG00000238860 |
. |
501873 |
0.0 |
| chr11_18414110_18414261 | 0.21 |
LDHA |
lactate dehydrogenase A |
1750 |
0.24 |
| chr12_68383944_68384095 | 0.21 |
IFNG-AS1 |
IFNG antisense RNA 1 |
710 |
0.81 |
| chr11_112033943_112034120 | 0.21 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
766 |
0.47 |
| chr4_48240161_48240435 | 0.21 |
TEC |
tec protein tyrosine kinase |
31583 |
0.18 |
| chr18_2848081_2848464 | 0.21 |
EMILIN2 |
elastin microfibril interfacer 2 |
1244 |
0.42 |
| chr1_185688405_185688780 | 0.21 |
HMCN1 |
hemicentin 1 |
15091 |
0.26 |
| chr4_70617052_70617203 | 0.21 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
9189 |
0.25 |
| chr19_14224361_14224512 | 0.21 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
564 |
0.58 |
| chr10_4284322_4284712 | 0.21 |
ENSG00000207124 |
. |
272627 |
0.02 |
| chr11_133666002_133666153 | 0.21 |
SPATA19 |
spermatogenesis associated 19 |
49319 |
0.16 |
| chr16_3309300_3309451 | 0.21 |
MEFV |
Mediterranean fever |
2748 |
0.14 |
| chr10_73585587_73585738 | 0.21 |
ENSG00000238446 |
. |
18889 |
0.16 |
| chr14_73928931_73929214 | 0.21 |
ENSG00000251393 |
. |
57 |
0.97 |
| chr3_134050593_134051228 | 0.21 |
AMOTL2 |
angiomotin like 2 |
39844 |
0.16 |
| chr14_65104571_65104722 | 0.21 |
RP11-973N13.3 |
|
41392 |
0.13 |
| chr17_49163006_49163241 | 0.21 |
ENSG00000238815 |
. |
12516 |
0.16 |
| chr9_130399731_130400130 | 0.21 |
STXBP1 |
syntaxin binding protein 1 |
25362 |
0.12 |
| chr2_51447556_51447707 | 0.20 |
NRXN1 |
neurexin 1 |
187957 |
0.03 |
| chr21_18348624_18348775 | 0.20 |
ENSG00000239023 |
. |
257382 |
0.02 |
| chr6_137555835_137556144 | 0.20 |
IFNGR1 |
interferon gamma receptor 1 |
15403 |
0.25 |
| chr2_40907638_40908036 | 0.20 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
69644 |
0.14 |
| chr1_234737377_234737528 | 0.20 |
RP4-781K5.2 |
|
5302 |
0.2 |
| chr15_90709177_90709401 | 0.20 |
ENSG00000240470 |
. |
176 |
0.92 |
| chr6_37093616_37093837 | 0.20 |
PIM1 |
pim-1 oncogene |
44253 |
0.13 |
| chr8_120221581_120221782 | 0.20 |
MAL2 |
mal, T-cell differentiation protein 2 (gene/pseudogene) |
1071 |
0.66 |
| chr6_144357186_144358268 | 0.20 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
28008 |
0.21 |
| chr5_88122444_88122620 | 0.20 |
MEF2C |
myocyte enhancer factor 2C |
443 |
0.88 |
| chr3_171788685_171788836 | 0.20 |
FNDC3B |
fibronectin type III domain containing 3B |
24057 |
0.25 |
| chr1_233393657_233393808 | 0.20 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
30717 |
0.22 |
| chr5_49738485_49738636 | 0.20 |
EMB |
embigin |
1359 |
0.62 |
| chr22_36231755_36232301 | 0.20 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
4237 |
0.32 |
| chr3_151608868_151609019 | 0.20 |
SUCNR1 |
succinate receptor 1 |
17512 |
0.21 |
| chr11_25264958_25265109 | 0.20 |
ENSG00000265837 |
. |
293791 |
0.01 |
| chr15_94796649_94796923 | 0.20 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
22019 |
0.29 |
| chr13_110746963_110747216 | 0.20 |
ENSG00000265885 |
. |
3409 |
0.37 |
| chr1_190210847_190210998 | 0.20 |
RP11-547I7.1 |
|
23106 |
0.29 |
| chr1_206553298_206553661 | 0.20 |
SRGAP2-AS1 |
SRGAP2 antisense RNA 1 |
1475 |
0.4 |
| chr1_40400191_40400342 | 0.20 |
ENSG00000207356 |
. |
10404 |
0.13 |
| chr13_49788937_49789202 | 0.20 |
MLNR |
motilin receptor |
5405 |
0.25 |
| chr12_113900471_113900622 | 0.20 |
RP11-82C23.2 |
|
9262 |
0.16 |
| chr14_107075724_107075875 | 0.20 |
IGHV3-57 |
immunoglobulin heavy variable 3-57 (pseudogene) |
613 |
0.3 |
| chr22_30591687_30592196 | 0.19 |
RP3-438O4.4 |
|
11157 |
0.14 |
| chr10_99444807_99445034 | 0.19 |
AVPI1 |
arginine vasopressin-induced 1 |
2160 |
0.25 |
| chr4_95489578_95489729 | 0.19 |
PDLIM5 |
PDZ and LIM domain 5 |
44777 |
0.21 |
| chr12_6495821_6496120 | 0.19 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
1685 |
0.24 |
| chr6_81964027_81964178 | 0.19 |
RP1-300G12.2 |
|
280528 |
0.01 |
| chr15_101273643_101273824 | 0.19 |
ENSG00000212306 |
. |
95165 |
0.07 |
| chr2_143904464_143904615 | 0.19 |
ARHGAP15 |
Rho GTPase activating protein 15 |
17656 |
0.23 |
| chr2_145216263_145216593 | 0.19 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
28291 |
0.23 |
| chr3_152884468_152884748 | 0.19 |
RAP2B |
RAP2B, member of RAS oncogene family |
4579 |
0.23 |
| chr2_10520029_10520465 | 0.19 |
HPCAL1 |
hippocalcin-like 1 |
39900 |
0.14 |
| chr4_30727938_30728161 | 0.19 |
PCDH7 |
protocadherin 7 |
4072 |
0.35 |
| chr2_196281841_196281992 | 0.19 |
ENSG00000202206 |
. |
96844 |
0.09 |
| chr5_34001934_34002085 | 0.19 |
AMACR |
alpha-methylacyl-CoA racemase |
6124 |
0.2 |
| chr7_129493207_129493836 | 0.19 |
ENSG00000207691 |
. |
78667 |
0.06 |
| chr12_70132575_70133879 | 0.19 |
RAB3IP |
RAB3A interacting protein |
47 |
0.95 |
| chr16_53241926_53242320 | 0.19 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
222 |
0.94 |
| chr11_27416621_27416772 | 0.19 |
CCDC34 |
coiled-coil domain containing 34 |
31281 |
0.18 |
| chr3_36937026_36937306 | 0.19 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
36792 |
0.17 |
| chr10_26779779_26779930 | 0.19 |
ENSG00000199733 |
. |
18664 |
0.24 |
| chr6_66730963_66731114 | 0.19 |
EYS |
eyes shut homolog (Drosophila) |
313920 |
0.01 |
| chr20_5260387_5260538 | 0.19 |
PROKR2 |
prokineticin receptor 2 |
34553 |
0.17 |
| chr1_64013670_64013821 | 0.19 |
DLEU2L |
deleted in lymphocytic leukemia 2-like |
843 |
0.52 |
| chr12_12491368_12491613 | 0.19 |
MANSC1 |
MANSC domain containing 1 |
118 |
0.96 |
| chr8_126232454_126232605 | 0.19 |
ENSG00000242170 |
. |
50317 |
0.15 |
| chr3_73633983_73634134 | 0.19 |
ENSG00000239119 |
. |
9095 |
0.22 |
| chr1_164529222_164529675 | 0.19 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
388 |
0.91 |
| chr6_130004791_130005136 | 0.19 |
ARHGAP18 |
Rho GTPase activating protein 18 |
26407 |
0.23 |
| chr1_33428156_33429049 | 0.19 |
RNF19B |
ring finger protein 19B |
1684 |
0.28 |
| chr3_60994671_60994822 | 0.19 |
ENSG00000212211 |
. |
152469 |
0.05 |
| chr5_97446035_97446186 | 0.19 |
ENSG00000223053 |
. |
528825 |
0.0 |
| chr1_36521425_36521576 | 0.19 |
TEKT2 |
tektin 2 (testicular) |
28176 |
0.13 |
| chr3_187607411_187607594 | 0.18 |
BCL6 |
B-cell CLL/lymphoma 6 |
143987 |
0.04 |
| chr1_224618149_224618304 | 0.18 |
WDR26 |
WD repeat domain 26 |
3581 |
0.24 |
| chr2_235275710_235275861 | 0.18 |
ARL4C |
ADP-ribosylation factor-like 4C |
129459 |
0.06 |
| chr6_82522367_82522541 | 0.18 |
ENSG00000206886 |
. |
48713 |
0.15 |
| chr21_30472506_30472657 | 0.18 |
MAP3K7CL |
MAP3K7 C-terminal like |
8006 |
0.14 |
| chr7_38670068_38671048 | 0.18 |
AMPH |
amphiphysin |
462 |
0.87 |
| chr15_58622989_58623389 | 0.18 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
51727 |
0.15 |
| chr4_100737483_100737634 | 0.18 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
432 |
0.88 |
| chr5_147566124_147566390 | 0.18 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
16100 |
0.18 |
| chr19_30714107_30714258 | 0.18 |
ZNF536 |
zinc finger protein 536 |
5239 |
0.26 |
| chrX_20413264_20413415 | 0.18 |
ENSG00000252978 |
. |
56887 |
0.16 |
| chr3_75291615_75291766 | 0.18 |
ENSG00000266780 |
. |
28063 |
0.21 |
| chr15_47476270_47477144 | 0.18 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
409 |
0.87 |
| chr3_43591104_43591255 | 0.18 |
ANO10 |
anoctamin 10 |
5724 |
0.29 |
| chr1_207509019_207509170 | 0.18 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
13958 |
0.26 |
| chr18_74205501_74206173 | 0.18 |
ZNF516 |
zinc finger protein 516 |
1309 |
0.32 |
| chrX_68063428_68063579 | 0.18 |
EFNB1 |
ephrin-B1 |
14663 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.1 | 0.7 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.1 | 0.3 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.1 | 0.3 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.3 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.3 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.0 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0010894 | negative regulation of steroid biosynthetic process(GO:0010894) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.0 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.0 | 0.1 | GO:0055057 | neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.0 | 0.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.0 | GO:2000649 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.5 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |