Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
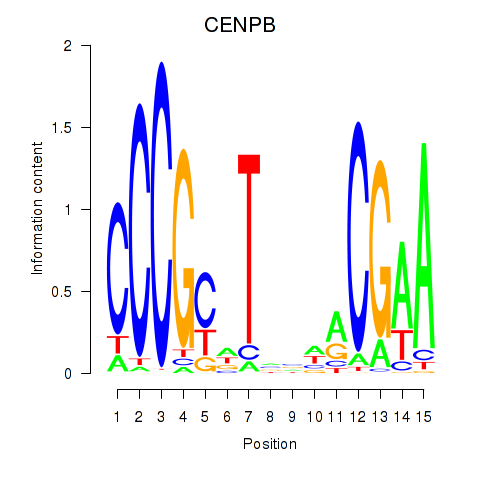
Results for CENPB
Z-value: 1.80
Transcription factors associated with CENPB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CENPB
|
ENSG00000125817.7 | centromere protein B |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_3766791_3767143 | CENPB | 370 | 0.599371 | 0.69 | 3.8e-02 | Click! |
| chr20_3764841_3764992 | CENPB | 2421 | 0.161262 | 0.44 | 2.4e-01 | Click! |
| chr20_3766238_3766588 | CENPB | 924 | 0.348276 | 0.39 | 3.0e-01 | Click! |
| chr20_3765290_3765684 | CENPB | 1850 | 0.197140 | 0.37 | 3.3e-01 | Click! |
| chr20_3765918_3766170 | CENPB | 1293 | 0.264619 | 0.36 | 3.4e-01 | Click! |
Activity of the CENPB motif across conditions
Conditions sorted by the z-value of the CENPB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_83378387_83378599 | 0.72 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
38 |
0.96 |
| chr9_116102276_116102560 | 0.67 |
WDR31 |
WD repeat domain 31 |
116 |
0.95 |
| chr5_60921640_60922072 | 0.66 |
C5orf64 |
chromosome 5 open reading frame 64 |
11780 |
0.22 |
| chr2_204735072_204735223 | 0.57 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
192 |
0.96 |
| chr4_42401015_42401166 | 0.55 |
SHISA3 |
shisa family member 3 |
1234 |
0.51 |
| chr20_52790515_52790830 | 0.54 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
160 |
0.97 |
| chr3_38496116_38496319 | 0.54 |
ACVR2B-AS1 |
ACVR2B antisense RNA 1 |
94 |
0.93 |
| chr8_130838389_130838591 | 0.52 |
RP11-473O4.5 |
|
6057 |
0.23 |
| chr10_89621181_89621332 | 0.51 |
PTEN |
phosphatase and tensin homolog |
1614 |
0.26 |
| chr11_66233385_66233536 | 0.50 |
PELI3 |
pellino E3 ubiquitin protein ligase family member 3 |
756 |
0.42 |
| chr19_17008131_17008363 | 0.48 |
CPAMD8 |
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
1221 |
0.33 |
| chr2_20251239_20251427 | 0.46 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
456 |
0.53 |
| chr6_1885704_1885932 | 0.46 |
FOXC1 |
forkhead box C1 |
275137 |
0.01 |
| chr22_31502736_31502961 | 0.46 |
SELM |
Selenoprotein M |
706 |
0.53 |
| chr17_59285555_59285706 | 0.46 |
RP11-136H19.1 |
|
72305 |
0.1 |
| chr2_233791696_233791935 | 0.45 |
NGEF |
neuronal guanine nucleotide exchange factor |
1049 |
0.54 |
| chr2_33172704_33172929 | 0.45 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
447 |
0.82 |
| chr1_85724953_85725320 | 0.45 |
C1orf52 |
chromosome 1 open reading frame 52 |
180 |
0.92 |
| chr21_45661777_45662023 | 0.45 |
ICOSLG |
inducible T-cell co-stimulator ligand |
1051 |
0.37 |
| chr10_104194159_104194310 | 0.44 |
CUEDC2 |
CUE domain containing 2 |
1816 |
0.17 |
| chr22_16192555_16192736 | 0.44 |
LL22NC03-N64E9.1 |
|
11641 |
0.18 |
| chr17_62658263_62658426 | 0.43 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
158 |
0.96 |
| chr9_15511271_15511422 | 0.43 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
329 |
0.87 |
| chr1_151763327_151763716 | 0.43 |
RP11-98D18.9 |
|
203 |
0.63 |
| chr1_1135870_1136021 | 0.43 |
TNFRSF18 |
tumor necrosis factor receptor superfamily, member 18 |
5115 |
0.08 |
| chr2_238876111_238876470 | 0.42 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
566 |
0.79 |
| chr3_58224088_58224320 | 0.42 |
ABHD6 |
abhydrolase domain containing 6 |
55 |
0.98 |
| chr3_57542507_57542919 | 0.42 |
PDE12 |
phosphodiesterase 12 |
709 |
0.52 |
| chr16_50581926_50582116 | 0.42 |
NKD1 |
naked cuticle homolog 1 (Drosophila) |
220 |
0.93 |
| chr5_95157288_95157503 | 0.41 |
GLRX |
glutaredoxin (thioltransferase) |
1020 |
0.47 |
| chr16_2827744_2828043 | 0.41 |
TCEB2 |
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
595 |
0.5 |
| chr10_179263_179512 | 0.40 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
1018 |
0.6 |
| chr20_61425995_61426159 | 0.40 |
MRGBP |
MRG/MORF4L binding protein |
1728 |
0.23 |
| chr1_168329622_168329773 | 0.40 |
ENSG00000207974 |
. |
15065 |
0.23 |
| chr3_152878700_152878913 | 0.40 |
RP11-529G21.2 |
|
758 |
0.56 |
| chr11_65222025_65222643 | 0.40 |
AP000769.1 |
Uncharacterized protein |
394 |
0.78 |
| chr5_176023977_176024277 | 0.40 |
GPRIN1 |
G protein regulated inducer of neurite outgrowth 1 |
13007 |
0.11 |
| chr9_136932210_136932418 | 0.39 |
BRD3 |
bromodomain containing 3 |
778 |
0.61 |
| chr13_107519622_107519843 | 0.39 |
ENSG00000239050 |
. |
107642 |
0.08 |
| chr1_202996029_202996239 | 0.39 |
PPFIA4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
508 |
0.72 |
| chr17_2907702_2908042 | 0.39 |
CTD-3060P21.1 |
|
38683 |
0.15 |
| chr5_34915226_34915588 | 0.39 |
BRIX1 |
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
74 |
0.96 |
| chr12_110905736_110905986 | 0.39 |
GPN3 |
GPN-loop GTPase 3 |
208 |
0.6 |
| chr8_33330699_33330850 | 0.38 |
FUT10 |
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
110 |
0.96 |
| chr11_62599289_62599501 | 0.38 |
STX5 |
syntaxin 5 |
118 |
0.68 |
| chr17_66288077_66288331 | 0.38 |
ARSG |
arylsulfatase G |
545 |
0.56 |
| chr18_268178_268573 | 0.38 |
THOC1 |
THO complex 1 |
348 |
0.92 |
| chr17_35849503_35849741 | 0.38 |
DUSP14 |
dual specificity phosphatase 14 |
315 |
0.9 |
| chr11_59523028_59523239 | 0.38 |
STX3 |
syntaxin 3 |
208 |
0.82 |
| chr7_65338891_65339374 | 0.38 |
VKORC1L1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
776 |
0.65 |
| chr5_140997244_140997566 | 0.38 |
AC008781.7 |
|
576 |
0.53 |
| chr3_72788121_72788360 | 0.38 |
ENSG00000222838 |
. |
47278 |
0.17 |
| chr17_48227691_48228226 | 0.38 |
PPP1R9B |
protein phosphatase 1, regulatory subunit 9B |
81 |
0.94 |
| chrY_2709555_2710183 | 0.37 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
92 |
0.98 |
| chr17_74235938_74236314 | 0.37 |
RNF157 |
ring finger protein 157 |
256 |
0.89 |
| chr1_212965305_212965712 | 0.37 |
TATDN3 |
TatD DNase domain containing 3 |
242 |
0.64 |
| chr15_63414046_63414224 | 0.36 |
LACTB |
lactamase, beta |
103 |
0.97 |
| chrX_90690150_90690364 | 0.36 |
PABPC5-AS1 |
PABPC5 antisense RNA 1 |
259 |
0.68 |
| chr19_52800063_52800231 | 0.36 |
ZNF480 |
zinc finger protein 480 |
283 |
0.77 |
| chr9_43134561_43134712 | 0.36 |
AL513478.1 |
Uncharacterized protein |
459 |
0.7 |
| chr15_29131227_29131499 | 0.36 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
54 |
0.98 |
| chr1_51769559_51769817 | 0.36 |
TTC39A |
tetratricopeptide repeat domain 39A |
6026 |
0.17 |
| chr9_71737251_71737695 | 0.36 |
TJP2 |
tight junction protein 2 |
1249 |
0.57 |
| chr12_49351468_49351675 | 0.35 |
ARF3 |
ADP-ribosylation factor 3 |
237 |
0.61 |
| chr17_73285759_73286238 | 0.35 |
SLC25A19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
407 |
0.72 |
| chr1_35545080_35545471 | 0.35 |
ZMYM1 |
zinc finger, MYM-type 1 |
283 |
0.91 |
| chr20_305267_305844 | 0.35 |
RP5-1103G7.4 |
|
323 |
0.68 |
| chr11_134201877_134202251 | 0.35 |
GLB1L2 |
galactosidase, beta 1-like 2 |
108 |
0.97 |
| chr9_133538924_133539075 | 0.35 |
PRDM12 |
PR domain containing 12 |
982 |
0.58 |
| chr16_67193901_67194322 | 0.35 |
TRADD |
TNFRSF1A-associated via death domain |
90 |
0.63 |
| chr2_204572665_204572956 | 0.35 |
CD28 |
CD28 molecule |
1394 |
0.52 |
| chr3_39448544_39448796 | 0.35 |
RPSA |
ribosomal protein SA |
444 |
0.68 |
| chr18_46477183_46477549 | 0.35 |
SMAD7 |
SMAD family member 7 |
285 |
0.93 |
| chr15_85113367_85113660 | 0.35 |
UBE2Q2P1 |
ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 1 |
418 |
0.57 |
| chr1_28833003_28833253 | 0.35 |
RCC1 |
regulator of chromosome condensation 1 |
559 |
0.45 |
| chr1_38510021_38510235 | 0.34 |
POU3F1 |
POU class 3 homeobox 1 |
2322 |
0.24 |
| chr4_1107333_1107580 | 0.34 |
RP11-20I20.2 |
|
29 |
0.58 |
| chr17_41440194_41440345 | 0.34 |
ENSG00000188825 |
. |
24516 |
0.11 |
| chr17_66959265_66959515 | 0.34 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
7898 |
0.24 |
| chr9_139334453_139334604 | 0.34 |
INPP5E |
inositol polyphosphate-5-phosphatase, 72 kDa |
254 |
0.85 |
| chr1_155658197_155658425 | 0.34 |
YY1AP1 |
YY1 associated protein 1 |
45 |
0.9 |
| chr14_99947789_99947995 | 0.34 |
CCNK |
cyclin K |
130 |
0.88 |
| chr11_7041299_7041574 | 0.33 |
ZNF214 |
zinc finger protein 214 |
30 |
0.76 |
| chr11_82612799_82613018 | 0.33 |
C11orf82 |
chromosome 11 open reading frame 82 |
124 |
0.5 |
| chr4_166754962_166755113 | 0.33 |
TLL1 |
tolloid-like 1 |
39373 |
0.22 |
| chr8_114446989_114447172 | 0.33 |
CSMD3 |
CUB and Sushi multiple domains 3 |
2032 |
0.42 |
| chr19_17136985_17137234 | 0.33 |
CPAMD8 |
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
516 |
0.7 |
| chr13_100068739_100068890 | 0.33 |
ENSG00000266207 |
. |
36909 |
0.15 |
| chr4_177116463_177116679 | 0.33 |
SPATA4 |
spermatogenesis associated 4 |
251 |
0.93 |
| chr7_21985259_21985518 | 0.33 |
CDCA7L |
cell division cycle associated 7-like |
108 |
0.98 |
| chr11_66725086_66725423 | 0.33 |
PC |
pyruvate carboxylase |
593 |
0.64 |
| chr2_87798273_87798528 | 0.33 |
RP11-1399P15.1 |
|
20847 |
0.27 |
| chr1_249133271_249133538 | 0.33 |
ZNF672 |
zinc finger protein 672 |
51 |
0.96 |
| chr15_26095105_26095679 | 0.33 |
ENSG00000266517 |
. |
1420 |
0.42 |
| chr2_136633081_136633631 | 0.33 |
MCM6 |
minichromosome maintenance complex component 6 |
640 |
0.71 |
| chr14_100784490_100784674 | 0.33 |
ENSG00000243976 |
. |
1125 |
0.26 |
| chr11_18416232_18416740 | 0.33 |
LDHA |
lactate dehydrogenase A |
235 |
0.88 |
| chr2_136875892_136876111 | 0.33 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
266 |
0.95 |
| chr1_116917904_116918072 | 0.33 |
ATP1A1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
1499 |
0.36 |
| chr22_40406174_40406325 | 0.32 |
FAM83F |
family with sequence similarity 83, member F |
557 |
0.75 |
| chr16_87526842_87527034 | 0.32 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
1287 |
0.49 |
| chr12_8113260_8113485 | 0.32 |
ENSG00000206636 |
. |
15483 |
0.14 |
| chr8_37605644_37605880 | 0.32 |
ERLIN2 |
ER lipid raft associated 2 |
10678 |
0.12 |
| chr11_843458_843641 | 0.32 |
TSPAN4 |
tetraspanin 4 |
210 |
0.78 |
| chr19_58898704_58898972 | 0.32 |
RPS5 |
ribosomal protein S5 |
182 |
0.75 |
| chr17_38716835_38717173 | 0.32 |
CCR7 |
chemokine (C-C motif) receptor 7 |
261 |
0.91 |
| chr1_39499733_39499961 | 0.31 |
NDUFS5 |
NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) |
7857 |
0.18 |
| chr14_74036748_74037002 | 0.31 |
ACOT2 |
acyl-CoA thioesterase 2 |
1112 |
0.35 |
| chr6_139094656_139094848 | 0.31 |
CCDC28A |
coiled-coil domain containing 28A |
95 |
0.97 |
| chr2_241500266_241500461 | 0.31 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
120 |
0.87 |
| chr17_14208922_14209145 | 0.31 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
4633 |
0.3 |
| chr14_50329070_50329296 | 0.31 |
ENSG00000265150 |
. |
384 |
0.75 |
| chr11_18719924_18720222 | 0.31 |
TMEM86A |
transmembrane protein 86A |
265 |
0.89 |
| chr2_220094595_220095282 | 0.31 |
ANKZF1 |
ankyrin repeat and zinc finger domain containing 1 |
257 |
0.63 |
| chr11_9635266_9635604 | 0.31 |
WEE1 |
WEE1 G2 checkpoint kinase |
27174 |
0.13 |
| chr1_26606672_26606961 | 0.31 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
203 |
0.9 |
| chr21_40211978_40212129 | 0.31 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
30493 |
0.21 |
| chr6_37786875_37787134 | 0.30 |
RP3-441A12.1 |
|
10 |
0.8 |
| chr17_8089749_8090065 | 0.30 |
ENSG00000266638 |
. |
586 |
0.5 |
| chr16_30968327_30968483 | 0.30 |
SETD1A |
SET domain containing 1A |
210 |
0.85 |
| chr7_151000467_151000632 | 0.30 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
25567 |
0.11 |
| chr10_124134109_124134260 | 0.30 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
28 |
0.98 |
| chr18_31158525_31158704 | 0.30 |
ASXL3 |
additional sex combs like 3 (Drosophila) |
9 |
0.99 |
| chr1_2162419_2162570 | 0.30 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
2360 |
0.2 |
| chr4_82136118_82136337 | 0.29 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
9 |
0.99 |
| chr14_22949288_22949494 | 0.29 |
ENSG00000251002 |
. |
887 |
0.35 |
| chr10_43859218_43859369 | 0.29 |
FXYD4 |
FXYD domain containing ion transport regulator 4 |
7797 |
0.17 |
| chr19_13136767_13137028 | 0.29 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
1086 |
0.34 |
| chr5_137224846_137225023 | 0.29 |
RP11-381K20.2 |
|
72 |
0.67 |
| chr12_864686_864837 | 0.29 |
WNK1 |
WNK lysine deficient protein kinase 1 |
2029 |
0.35 |
| chr9_2157815_2158699 | 0.28 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
199 |
0.95 |
| chr17_60142459_60142618 | 0.28 |
MED13 |
mediator complex subunit 13 |
105 |
0.97 |
| chr3_149531082_149531931 | 0.28 |
RNF13 |
ring finger protein 13 |
101 |
0.97 |
| chr20_39311567_39311981 | 0.28 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
5724 |
0.34 |
| chr16_56709880_56710031 | 0.28 |
MT1H |
metallothionein 1H |
6203 |
0.08 |
| chr1_154581072_154581232 | 0.28 |
ADAR |
adenosine deaminase, RNA-specific |
470 |
0.73 |
| chr10_126480570_126481048 | 0.28 |
METTL10 |
methyltransferase like 10 |
370 |
0.83 |
| chr11_16759613_16759958 | 0.28 |
C11orf58 |
chromosome 11 open reading frame 58 |
163 |
0.97 |
| chr6_144982221_144982372 | 0.28 |
UTRN |
utrophin |
1316 |
0.63 |
| chr20_47796571_47796722 | 0.28 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
5846 |
0.21 |
| chr6_106536140_106536635 | 0.28 |
PRDM1 |
PR domain containing 1, with ZNF domain |
932 |
0.64 |
| chr13_97874593_97875664 | 0.28 |
MBNL2 |
muscleblind-like splicing regulator 2 |
519 |
0.87 |
| chr17_81060853_81061004 | 0.28 |
METRNL |
meteorin, glial cell differentiation regulator-like |
8934 |
0.25 |
| chr9_134152374_134152613 | 0.28 |
FAM78A |
family with sequence similarity 78, member A |
559 |
0.75 |
| chr10_21822138_21822619 | 0.27 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
716 |
0.59 |
| chr5_90677658_90677809 | 0.27 |
ARRDC3 |
arrestin domain containing 3 |
1443 |
0.51 |
| chr1_23894723_23894953 | 0.27 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
8553 |
0.18 |
| chr8_145980713_145981003 | 0.27 |
ZNF251 |
zinc finger protein 251 |
32 |
0.96 |
| chr4_56914817_56915143 | 0.27 |
ENSG00000200741 |
. |
48576 |
0.15 |
| chr16_25123142_25123518 | 0.27 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
77 |
0.98 |
| chr5_171434068_171434407 | 0.27 |
FBXW11 |
F-box and WD repeat domain containing 11 |
360 |
0.9 |
| chr13_109148449_109148673 | 0.27 |
MYO16 |
myosin XVI |
99939 |
0.08 |
| chr5_68514057_68514286 | 0.26 |
MRPS36 |
mitochondrial ribosomal protein S36 |
512 |
0.71 |
| chr8_1772294_1772657 | 0.26 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
294 |
0.91 |
| chr1_155145539_155145727 | 0.26 |
KRTCAP2 |
keratinocyte associated protein 2 |
176 |
0.54 |
| chr10_131264926_131265255 | 0.26 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
358 |
0.9 |
| chr12_32292111_32292535 | 0.26 |
RP11-843B15.2 |
|
31861 |
0.19 |
| chr1_15482935_15483201 | 0.26 |
TMEM51 |
transmembrane protein 51 |
2783 |
0.3 |
| chr9_35072054_35072271 | 0.26 |
VCP |
valosin containing protein |
151 |
0.91 |
| chr10_131762270_131762646 | 0.26 |
EBF3 |
early B-cell factor 3 |
353 |
0.93 |
| chr15_63449409_63449703 | 0.26 |
RPS27L |
ribosomal protein S27-like |
76 |
0.97 |
| chr10_126490181_126490332 | 0.26 |
FAM175B |
family with sequence similarity 175, member B |
98 |
0.96 |
| chr1_156405353_156405868 | 0.26 |
C1orf61 |
chromosome 1 open reading frame 61 |
6395 |
0.13 |
| chr1_155532313_155532497 | 0.26 |
ASH1L |
ash1 (absent, small, or homeotic)-like (Drosophila) |
79 |
0.89 |
| chr14_88459061_88459223 | 0.26 |
GALC |
galactosylceramidase |
40 |
0.97 |
| chr1_38757878_38758029 | 0.26 |
ENSG00000200796 |
. |
104378 |
0.07 |
| chr7_138916860_138917323 | 0.26 |
UBN2 |
ubinuclein 2 |
860 |
0.66 |
| chr5_108745263_108745487 | 0.26 |
PJA2 |
praja ring finger 2, E3 ubiquitin protein ligase |
320 |
0.95 |
| chr12_56522678_56523035 | 0.26 |
RP11-603J24.5 |
|
218 |
0.74 |
| chr14_71338943_71339141 | 0.26 |
PCNX |
pecanex homolog (Drosophila) |
35080 |
0.2 |
| chrX_153084272_153084494 | 0.26 |
PDZD4 |
PDZ domain containing 4 |
10968 |
0.09 |
| chr12_2377393_2377544 | 0.26 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
1474 |
0.5 |
| chr14_90320573_90320724 | 0.26 |
EFCAB11 |
EF-hand calcium binding domain 11 |
100219 |
0.07 |
| chr2_241374046_241374232 | 0.26 |
GPC1 |
glypican 1 |
949 |
0.53 |
| chr17_48906674_48906825 | 0.26 |
WFIKKN2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
5262 |
0.17 |
| chr11_61197616_61198048 | 0.26 |
SDHAF2 |
succinate dehydrogenase complex assembly factor 2 |
216 |
0.45 |
| chr11_58346016_58346264 | 0.26 |
ZFP91 |
ZFP91 zinc finger protein |
444 |
0.45 |
| chr13_46038649_46038966 | 0.26 |
COG3 |
component of oligomeric golgi complex 3 |
253 |
0.93 |
| chr5_134374502_134374924 | 0.26 |
PITX1 |
paired-like homeodomain 1 |
4210 |
0.23 |
| chr11_65430663_65431111 | 0.26 |
RELA |
v-rel avian reticuloendotheliosis viral oncogene homolog A |
322 |
0.76 |
| chr19_12917473_12917666 | 0.26 |
RNASEH2A |
ribonuclease H2, subunit A |
175 |
0.85 |
| chr6_13486568_13486897 | 0.25 |
GFOD1-AS1 |
GFOD1 antisense RNA 1 |
206 |
0.6 |
| chr11_64637981_64638132 | 0.25 |
EHD1 |
EH-domain containing 1 |
5082 |
0.11 |
| chr20_34542852_34543053 | 0.25 |
SCAND1 |
SCAN domain containing 1 |
404 |
0.81 |
| chr17_74383852_74384098 | 0.25 |
SPHK1 |
sphingosine kinase 1 |
2602 |
0.17 |
| chr11_58978519_58978670 | 0.25 |
MPEG1 |
macrophage expressed 1 |
1830 |
0.29 |
| chr12_69005315_69005553 | 0.25 |
RAP1B |
RAP1B, member of RAS oncogene family |
629 |
0.75 |
| chr1_170655624_170655775 | 0.25 |
PRRX1 |
paired related homeobox 1 |
22621 |
0.26 |
| chr15_59225515_59225703 | 0.25 |
SLTM |
SAFB-like, transcription modulator |
164 |
0.95 |
| chr6_24910724_24911222 | 0.25 |
FAM65B |
family with sequence similarity 65, member B |
222 |
0.95 |
| chr7_855243_855541 | 0.25 |
SUN1 |
Sad1 and UNC84 domain containing 1 |
136 |
0.96 |
| chr15_83377850_83378150 | 0.25 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
531 |
0.69 |
| chr17_17206244_17206395 | 0.25 |
NT5M |
5',3'-nucleotidase, mitochondrial |
330 |
0.87 |
| chr15_55880849_55881113 | 0.25 |
PYGO1 |
pygopus family PHD finger 1 |
164 |
0.97 |
| chr10_13342075_13342296 | 0.25 |
PHYH |
phytanoyl-CoA 2-hydroxylase |
84 |
0.97 |
| chr2_87883698_87883984 | 0.25 |
ENSG00000265507 |
. |
45433 |
0.18 |
| chr3_195906840_195907022 | 0.25 |
ENSG00000222335 |
. |
22289 |
0.13 |
| chr22_38349726_38350087 | 0.25 |
POLR2F |
polymerase (RNA) II (DNA directed) polypeptide F |
172 |
0.56 |
| chr22_42486316_42486738 | 0.25 |
NDUFA6 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
256 |
0.5 |
| chr17_41437916_41438239 | 0.25 |
ENSG00000188825 |
. |
26708 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.2 | 0.5 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.6 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.4 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.4 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.3 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.3 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.1 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.2 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.3 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.8 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.1 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.2 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.1 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.1 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0071027 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 0.3 | GO:0035872 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.4 | GO:0042634 | regulation of hair cycle(GO:0042634) |
| 0.1 | 0.2 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.2 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.1 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.2 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.1 | 0.2 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.2 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.3 | GO:0035510 | DNA dealkylation(GO:0035510) |
| 0.0 | 0.3 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.3 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.3 | GO:0045350 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.3 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.2 | GO:0055026 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0006862 | nucleotide transport(GO:0006862) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.2 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.3 | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process(GO:0009148) |
| 0.0 | 0.2 | GO:1901160 | serotonin metabolic process(GO:0042428) primary amino compound metabolic process(GO:1901160) |
| 0.0 | 0.0 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.0 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.4 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.0 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.0 | 0.1 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.8 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0033233 | regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.1 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.0 | GO:0060033 | Mullerian duct regression(GO:0001880) anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0050955 | sensory perception of temperature stimulus(GO:0050951) thermoception(GO:0050955) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.0 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0043501 | skeletal muscle adaptation(GO:0043501) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0051956 | negative regulation of amino acid transport(GO:0051956) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.0 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.0 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.0 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 1.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.0 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.1 | GO:0006184 | obsolete GTP catabolic process(GO:0006184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.1 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.3 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.9 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.0 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 1.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.3 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0030288 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.0 | GO:0043230 | extracellular organelle(GO:0043230) extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.2 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.1 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.3 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.3 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.7 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.5 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.6 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0015421 | oligopeptide-transporting ATPase activity(GO:0015421) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0034595 | phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.0 | 0.2 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.8 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.8 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 1.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.8 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |