Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
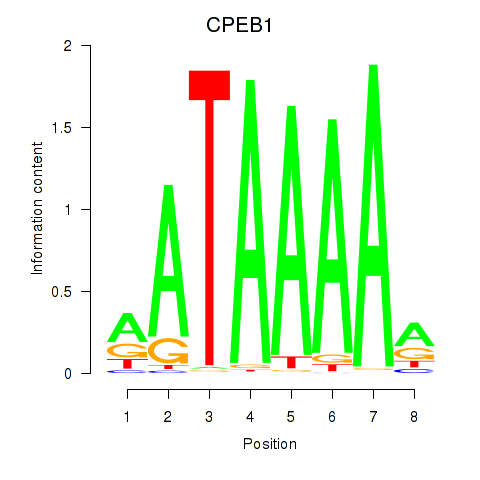
Results for CPEB1
Z-value: 1.45
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
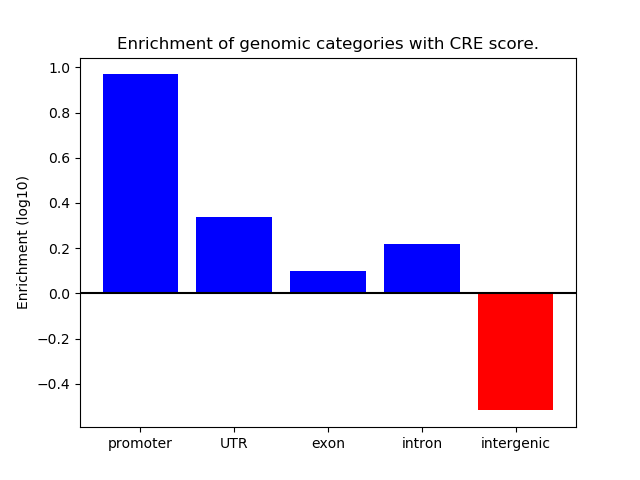
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_83240603_83240843 | CPEB1 | 105 | 0.950358 | -0.65 | 5.9e-02 | Click! |
| chr15_83239771_83240251 | CPEB1 | 498 | 0.712993 | -0.59 | 9.2e-02 | Click! |
| chr15_83240421_83240572 | CPEB1 | 13 | 0.965750 | -0.36 | 3.4e-01 | Click! |
| chr15_83316430_83317127 | CPEB1 | 50 | 0.760784 | 0.30 | 4.4e-01 | Click! |
| chr15_83316186_83316426 | CPEB1 | 50 | 0.716646 | 0.24 | 5.3e-01 | Click! |
Activity of the CPEB1 motif across conditions
Conditions sorted by the z-value of the CPEB1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
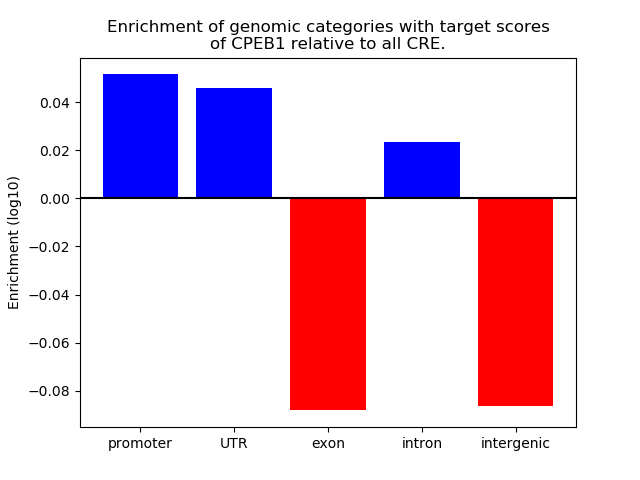
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_99726709_99727240 | 0.71 |
AL109767.1 |
|
2311 |
0.33 |
| chr3_112217277_112217447 | 0.66 |
BTLA |
B and T lymphocyte associated |
843 |
0.68 |
| chr14_91883398_91883667 | 0.55 |
CCDC88C |
coiled-coil domain containing 88C |
158 |
0.97 |
| chr2_197033343_197033494 | 0.52 |
STK17B |
serine/threonine kinase 17b |
2306 |
0.31 |
| chr15_80262648_80263745 | 0.48 |
BCL2A1 |
BCL2-related protein A1 |
315 |
0.89 |
| chr1_65428751_65428968 | 0.42 |
JAK1 |
Janus kinase 1 |
3328 |
0.31 |
| chr11_58340730_58341020 | 0.42 |
LPXN |
leupaxin |
2459 |
0.23 |
| chr3_18475636_18475787 | 0.42 |
SATB1 |
SATB homeobox 1 |
1034 |
0.57 |
| chr10_74008100_74008285 | 0.40 |
DDIT4 |
DNA-damage-inducible transcript 4 |
25486 |
0.12 |
| chr22_31686352_31686503 | 0.40 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
1954 |
0.19 |
| chr22_32858699_32858850 | 0.40 |
BPIFC |
BPI fold containing family C |
1655 |
0.35 |
| chr2_213977055_213977206 | 0.40 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
36223 |
0.21 |
| chr18_60874994_60875145 | 0.39 |
ENSG00000238988 |
. |
13171 |
0.23 |
| chr8_113941673_113941824 | 0.38 |
CSMD3 |
CUB and Sushi multiple domains 3 |
239477 |
0.02 |
| chr2_198160747_198160898 | 0.36 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
6421 |
0.18 |
| chrX_73071645_73072457 | 0.36 |
RP13-216E22.5 |
|
117269 |
0.06 |
| chr4_48135139_48135397 | 0.36 |
TXK |
TXK tyrosine kinase |
1005 |
0.5 |
| chr18_9107105_9107411 | 0.36 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
3178 |
0.21 |
| chr7_45075825_45075976 | 0.35 |
CCM2 |
cerebral cavernous malformation 2 |
8629 |
0.15 |
| chr1_198609469_198609717 | 0.34 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
1301 |
0.54 |
| chr6_12010262_12011396 | 0.34 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
1404 |
0.51 |
| chrX_64255707_64255858 | 0.34 |
ZC4H2 |
zinc finger, C4H2 domain containing |
1189 |
0.66 |
| chr5_162932121_162932408 | 0.34 |
MAT2B |
methionine adenosyltransferase II, beta |
290 |
0.89 |
| chr10_11252091_11252492 | 0.34 |
RP3-323N1.2 |
|
38952 |
0.17 |
| chr16_66864042_66864206 | 0.34 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
696 |
0.56 |
| chr14_52327497_52328482 | 0.34 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
53 |
0.98 |
| chr10_63753311_63753523 | 0.34 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
55553 |
0.14 |
| chr19_19475616_19475786 | 0.33 |
GATAD2A |
GATA zinc finger domain containing 2A |
20934 |
0.12 |
| chr14_22972981_22973242 | 0.33 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
16940 |
0.09 |
| chr11_5246641_5246814 | 0.32 |
ENSG00000221031 |
. |
858 |
0.32 |
| chr19_16438433_16439148 | 0.32 |
KLF2 |
Kruppel-like factor 2 |
3139 |
0.19 |
| chr1_198608588_198608957 | 0.32 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
480 |
0.86 |
| chr17_29639732_29640055 | 0.32 |
EVI2B |
ecotropic viral integration site 2B |
1209 |
0.36 |
| chr9_273119_273416 | 0.32 |
DOCK8 |
dedicator of cytokinesis 8 |
197 |
0.59 |
| chr12_6555099_6555250 | 0.32 |
CD27 |
CD27 molecule |
1141 |
0.3 |
| chrX_78201853_78202096 | 0.32 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
1056 |
0.69 |
| chr14_99734042_99734488 | 0.31 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
3300 |
0.26 |
| chr10_43843775_43843959 | 0.31 |
ENSG00000221468 |
. |
6633 |
0.19 |
| chr2_158299517_158299677 | 0.31 |
CYTIP |
cytohesin 1 interacting protein |
1057 |
0.48 |
| chr6_170862470_170862790 | 0.31 |
PSMB1 |
proteasome (prosome, macropain) subunit, beta type, 1 |
201 |
0.86 |
| chr6_24933660_24933811 | 0.30 |
FAM65B |
family with sequence similarity 65, member B |
2453 |
0.34 |
| chr1_173988808_173989095 | 0.30 |
RC3H1 |
ring finger and CCCH-type domains 1 |
2484 |
0.25 |
| chr6_106547367_106547905 | 0.30 |
RP1-134E15.3 |
|
379 |
0.74 |
| chr1_203735045_203735272 | 0.30 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
853 |
0.57 |
| chr3_151922606_151922776 | 0.30 |
MBNL1 |
muscleblind-like splicing regulator 1 |
63138 |
0.12 |
| chr11_6767215_6767876 | 0.30 |
OR2AG2 |
olfactory receptor, family 2, subfamily AG, member 2 |
22741 |
0.11 |
| chr5_57122892_57123043 | 0.29 |
ENSG00000266864 |
. |
45906 |
0.2 |
| chrX_153656709_153656889 | 0.29 |
ATP6AP1 |
ATPase, H+ transporting, lysosomal accessory protein 1 |
179 |
0.86 |
| chr14_22974481_22974956 | 0.29 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18547 |
0.09 |
| chr5_110562007_110562166 | 0.29 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
2302 |
0.35 |
| chr3_32993605_32993756 | 0.29 |
CCR4 |
chemokine (C-C motif) receptor 4 |
614 |
0.81 |
| chr18_24853185_24853336 | 0.29 |
CHST9 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
87979 |
0.11 |
| chr5_130882612_130883040 | 0.29 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
14100 |
0.29 |
| chr17_33859440_33859591 | 0.29 |
SLFN12L |
schlafen family member 12-like |
5365 |
0.12 |
| chr20_47436634_47436796 | 0.29 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
7705 |
0.28 |
| chr19_2084472_2084623 | 0.29 |
MOB3A |
MOB kinase activator 3A |
844 |
0.46 |
| chr13_52398191_52398536 | 0.28 |
RP11-327P2.5 |
|
19930 |
0.17 |
| chr13_31309749_31309959 | 0.28 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
209 |
0.96 |
| chr1_89735192_89735432 | 0.28 |
GBP5 |
guanylate binding protein 5 |
1234 |
0.47 |
| chr2_68962051_68962936 | 0.28 |
ARHGAP25 |
Rho GTPase activating protein 25 |
479 |
0.86 |
| chr19_42057151_42057302 | 0.28 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
1340 |
0.4 |
| chr19_18197219_18197405 | 0.28 |
IL12RB1 |
interleukin 12 receptor, beta 1 |
430 |
0.78 |
| chr9_95821637_95821823 | 0.28 |
SUSD3 |
sushi domain containing 3 |
669 |
0.69 |
| chr18_32609618_32610414 | 0.27 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
11308 |
0.29 |
| chr12_56754222_56754408 | 0.27 |
STAT2 |
signal transducer and activator of transcription 2, 113kDa |
405 |
0.67 |
| chr7_19955546_19955697 | 0.27 |
AC005062.2 |
|
52239 |
0.16 |
| chr3_151962256_151962407 | 0.27 |
MBNL1 |
muscleblind-like splicing regulator 1 |
23498 |
0.2 |
| chr19_42390602_42391041 | 0.27 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
2306 |
0.19 |
| chr11_82828119_82828301 | 0.27 |
PCF11 |
PCF11 cleavage and polyadenylation factor subunit |
39820 |
0.12 |
| chr15_34626880_34627184 | 0.27 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
2013 |
0.2 |
| chr1_179846681_179846874 | 0.27 |
TOR1AIP2 |
torsin A interacting protein 2 |
157 |
0.94 |
| chr2_204801338_204802214 | 0.27 |
ICOS |
inducible T-cell co-stimulator |
273 |
0.95 |
| chr19_6669496_6669799 | 0.27 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
481 |
0.75 |
| chr14_35763113_35763264 | 0.27 |
PSMA6 |
proteasome (prosome, macropain) subunit, alpha type, 6 |
1509 |
0.4 |
| chr6_112079133_112079284 | 0.26 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1109 |
0.63 |
| chr2_64368851_64369687 | 0.26 |
AC074289.1 |
|
1104 |
0.52 |
| chr22_37373947_37374259 | 0.26 |
LL22NC01-81G9.3 |
|
12814 |
0.11 |
| chr5_130602420_130602583 | 0.26 |
CDC42SE2 |
CDC42 small effector 2 |
2708 |
0.39 |
| chr17_66031093_66031463 | 0.26 |
KPNA2 |
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
357 |
0.87 |
| chr22_38788070_38788476 | 0.26 |
RP1-5O6.4 |
|
3411 |
0.16 |
| chr1_160614614_160614765 | 0.26 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
2122 |
0.27 |
| chr4_54232426_54232649 | 0.26 |
SCFD2 |
sec1 family domain containing 2 |
295 |
0.92 |
| chr20_21528926_21529077 | 0.26 |
NKX2-2 |
NK2 homeobox 2 |
34337 |
0.18 |
| chr20_35224303_35224506 | 0.26 |
RP5-977B1.11 |
|
8528 |
0.12 |
| chr6_90790037_90790282 | 0.26 |
ENSG00000222078 |
. |
78934 |
0.1 |
| chr3_46413606_46413757 | 0.26 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
480 |
0.77 |
| chr2_175459196_175459684 | 0.25 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
3053 |
0.24 |
| chr4_109086470_109087224 | 0.25 |
LEF1 |
lymphoid enhancer-binding factor 1 |
610 |
0.72 |
| chr1_156183089_156183279 | 0.25 |
PMF1-BGLAP |
PMF1-BGLAP readthrough |
384 |
0.43 |
| chrX_41195972_41196123 | 0.25 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
2203 |
0.27 |
| chr8_71287137_71287766 | 0.25 |
ENSG00000207036 |
. |
17869 |
0.21 |
| chr9_86536534_86536716 | 0.25 |
KIF27 |
kinesin family member 27 |
283 |
0.88 |
| chr14_23009644_23009795 | 0.25 |
TRAJ15 |
T cell receptor alpha joining 15 |
11139 |
0.1 |
| chr20_12390339_12390490 | 0.25 |
BTBD3 |
BTB (POZ) domain containing 3 |
490915 |
0.01 |
| chr5_110565458_110565614 | 0.25 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
5752 |
0.25 |
| chr8_29955696_29955905 | 0.25 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
2635 |
0.22 |
| chr16_28998125_28998276 | 0.25 |
LAT |
linker for activation of T cells |
667 |
0.47 |
| chr3_43330385_43330567 | 0.25 |
SNRK |
SNF related kinase |
2398 |
0.29 |
| chr11_128386110_128386459 | 0.25 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
5812 |
0.25 |
| chr2_202127394_202127951 | 0.24 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
993 |
0.56 |
| chr17_37932973_37933124 | 0.24 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1430 |
0.33 |
| chr17_29637048_29637296 | 0.24 |
EVI2B |
ecotropic viral integration site 2B |
3930 |
0.16 |
| chr1_158902534_158902685 | 0.24 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
1251 |
0.51 |
| chr12_109023643_109023867 | 0.24 |
RP11-689B22.2 |
|
1292 |
0.32 |
| chr16_68798920_68799109 | 0.24 |
ENSG00000200558 |
. |
22631 |
0.14 |
| chr12_52885749_52885900 | 0.24 |
KRT6A |
keratin 6A |
1217 |
0.31 |
| chr11_64547205_64547356 | 0.24 |
SF1 |
splicing factor 1 |
1022 |
0.39 |
| chr9_4285477_4285628 | 0.24 |
GLIS3 |
GLIS family zinc finger 3 |
12944 |
0.21 |
| chr1_234544831_234544982 | 0.24 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
35459 |
0.12 |
| chr12_104858566_104858717 | 0.24 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
7862 |
0.28 |
| chr2_237446552_237446703 | 0.24 |
ACKR3 |
atypical chemokine receptor 3 |
29803 |
0.19 |
| chr18_2662663_2662814 | 0.24 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
6852 |
0.17 |
| chr14_102290508_102290727 | 0.24 |
CTD-2017C7.2 |
|
13959 |
0.14 |
| chr11_128395074_128395489 | 0.24 |
RP11-1007G5.2 |
|
756 |
0.69 |
| chr2_143887894_143888150 | 0.24 |
ARHGAP15 |
Rho GTPase activating protein 15 |
1139 |
0.61 |
| chr11_109633690_109633841 | 0.24 |
RP11-708B6.2 |
|
179132 |
0.03 |
| chr4_70625524_70625675 | 0.24 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
717 |
0.76 |
| chr6_106500939_106501090 | 0.24 |
PRDM1 |
PR domain containing 1, with ZNF domain |
33181 |
0.19 |
| chr12_32114494_32114645 | 0.24 |
KIAA1551 |
KIAA1551 |
854 |
0.68 |
| chr2_208666293_208666444 | 0.23 |
FZD5 |
frizzled family receptor 5 |
32081 |
0.13 |
| chr7_150440682_150440833 | 0.23 |
GIMAP5 |
GTPase, IMAP family member 5 |
6321 |
0.17 |
| chr9_128592071_128592626 | 0.23 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
35202 |
0.22 |
| chrX_16120698_16120849 | 0.23 |
GRPR |
gastrin-releasing peptide receptor |
20906 |
0.21 |
| chr14_22739514_22739665 | 0.23 |
ENSG00000238634 |
. |
128702 |
0.04 |
| chr6_106549276_106549427 | 0.23 |
RP1-134E15.3 |
|
1336 |
0.46 |
| chr2_231304576_231304741 | 0.23 |
SP100 |
SP100 nuclear antigen |
22492 |
0.21 |
| chr22_36925707_36925858 | 0.23 |
EIF3D |
eukaryotic translation initiation factor 3, subunit D |
299 |
0.87 |
| chr15_57199386_57199736 | 0.23 |
ZNF280D |
zinc finger protein 280D |
11208 |
0.22 |
| chr1_161041599_161041750 | 0.23 |
ARHGAP30 |
Rho GTPase activating protein 30 |
1914 |
0.15 |
| chr2_198172097_198172605 | 0.23 |
ANKRD44 |
ankyrin repeat domain 44 |
3160 |
0.2 |
| chr11_80154667_80154818 | 0.23 |
ENSG00000200146 |
. |
84262 |
0.12 |
| chr6_112078365_112078719 | 0.23 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1775 |
0.47 |
| chr3_104938786_104938937 | 0.23 |
ALCAM |
activated leukocyte cell adhesion molecule |
146892 |
0.05 |
| chr2_143888217_143888368 | 0.23 |
ARHGAP15 |
Rho GTPase activating protein 15 |
1409 |
0.53 |
| chr6_106548995_106549243 | 0.23 |
RP1-134E15.3 |
|
1104 |
0.52 |
| chr19_42082862_42083083 | 0.23 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
371 |
0.84 |
| chr1_186291918_186292069 | 0.23 |
ENSG00000202025 |
. |
10933 |
0.17 |
| chr2_175498427_175499207 | 0.23 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
490 |
0.83 |
| chr2_143888975_143889165 | 0.23 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2187 |
0.4 |
| chr17_76439506_76439657 | 0.23 |
AC061992.1 |
Uncharacterized protein |
17172 |
0.14 |
| chr2_158290873_158291040 | 0.23 |
CYTIP |
cytohesin 1 interacting protein |
4970 |
0.24 |
| chr14_50364801_50365248 | 0.23 |
ENSG00000251929 |
. |
3644 |
0.15 |
| chr5_45041940_45042091 | 0.23 |
MRPS30 |
mitochondrial ribosomal protein S30 |
232988 |
0.02 |
| chr2_37828420_37828576 | 0.23 |
AC006369.2 |
|
1219 |
0.57 |
| chr13_41588793_41589349 | 0.23 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
4379 |
0.23 |
| chr5_110561342_110561544 | 0.23 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1659 |
0.44 |
| chr1_239884533_239884684 | 0.23 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
1765 |
0.38 |
| chr12_96896557_96896708 | 0.22 |
C12orf55 |
chromosome 12 open reading frame 55 |
13247 |
0.23 |
| chr15_79166100_79166403 | 0.22 |
MORF4L1 |
mortality factor 4 like 1 |
88 |
0.97 |
| chr3_39372377_39372528 | 0.22 |
CCR8 |
chemokine (C-C motif) receptor 8 |
1188 |
0.47 |
| chr14_99720527_99720678 | 0.22 |
AL109767.1 |
|
8683 |
0.22 |
| chr2_237446083_237446280 | 0.22 |
IQCA1 |
IQ motif containing with AAA domain 1 |
29996 |
0.19 |
| chr2_20549100_20549748 | 0.22 |
PUM2 |
pumilio RNA-binding family member 2 |
958 |
0.54 |
| chr13_99953472_99953749 | 0.22 |
GPR183 |
G protein-coupled receptor 183 |
6049 |
0.22 |
| chr14_38546459_38546610 | 0.22 |
CTD-2058B24.2 |
|
13829 |
0.25 |
| chr8_19354385_19354536 | 0.22 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
104916 |
0.08 |
| chr6_130535166_130535317 | 0.22 |
SAMD3 |
sterile alpha motif domain containing 3 |
1194 |
0.61 |
| chr15_68591045_68591196 | 0.22 |
FEM1B |
fem-1 homolog b (C. elegans) |
8570 |
0.21 |
| chr10_82228022_82228360 | 0.22 |
TSPAN14 |
tetraspanin 14 |
9133 |
0.21 |
| chr2_117455100_117455251 | 0.22 |
ENSG00000239185 |
. |
442176 |
0.01 |
| chr12_65063818_65064050 | 0.22 |
RP11-338E21.3 |
|
15135 |
0.13 |
| chr5_151907362_151907513 | 0.22 |
NMUR2 |
neuromedin U receptor 2 |
122597 |
0.07 |
| chr17_43297016_43297167 | 0.22 |
CTD-2020K17.1 |
|
608 |
0.54 |
| chr14_22978649_22978851 | 0.22 |
TRAJ15 |
T cell receptor alpha joining 15 |
19830 |
0.09 |
| chr10_62493530_62493681 | 0.22 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
357 |
0.92 |
| chr2_99327752_99327916 | 0.22 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
15325 |
0.22 |
| chr1_112167264_112167415 | 0.22 |
RAP1A |
RAP1A, member of RAS oncogene family |
4570 |
0.2 |
| chr13_42631023_42631174 | 0.22 |
DGKH |
diacylglycerol kinase, eta |
8209 |
0.22 |
| chr5_40410059_40410210 | 0.22 |
ENSG00000265615 |
. |
89714 |
0.09 |
| chr8_19613768_19613970 | 0.22 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
947 |
0.69 |
| chr7_140176949_140177100 | 0.22 |
MKRN1 |
makorin ring finger protein 1 |
1279 |
0.46 |
| chr17_43661192_43661756 | 0.21 |
RP11-798G7.4 |
|
25038 |
0.12 |
| chr8_82011756_82011907 | 0.21 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
12472 |
0.28 |
| chr5_133455200_133455351 | 0.21 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
3959 |
0.26 |
| chr2_136348761_136348912 | 0.21 |
R3HDM1 |
R3H domain containing 1 |
4829 |
0.25 |
| chr12_118796178_118796667 | 0.21 |
TAOK3 |
TAO kinase 3 |
488 |
0.85 |
| chr18_12775334_12775517 | 0.21 |
ENSG00000201466 |
. |
14709 |
0.18 |
| chr20_1470626_1470777 | 0.21 |
SIRPB2 |
signal-regulatory protein beta 2 |
1341 |
0.29 |
| chrX_135861825_135862156 | 0.21 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
2257 |
0.28 |
| chr17_73293586_73293836 | 0.21 |
SLC25A19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
8120 |
0.1 |
| chr2_3082956_3083107 | 0.21 |
ENSG00000238722 |
. |
3487 |
0.34 |
| chr19_48762542_48762841 | 0.21 |
CARD8 |
caspase recruitment domain family, member 8 |
3488 |
0.14 |
| chr3_46414243_46414394 | 0.21 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
157 |
0.95 |
| chr7_151622563_151622783 | 0.21 |
ENSG00000222941 |
. |
9261 |
0.14 |
| chr3_166970881_166971032 | 0.21 |
ZBBX |
zinc finger, B-box domain containing |
119820 |
0.06 |
| chr2_197028035_197028292 | 0.21 |
STK17B |
serine/threonine kinase 17b |
6792 |
0.21 |
| chr12_104856651_104856876 | 0.21 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
5984 |
0.3 |
| chr2_143889174_143889389 | 0.21 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2398 |
0.38 |
| chr17_37948516_37948667 | 0.21 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
14113 |
0.14 |
| chr1_203259093_203259633 | 0.21 |
BTG2 |
BTG family, member 2 |
15301 |
0.16 |
| chr5_39190229_39190380 | 0.21 |
FYB |
FYN binding protein |
12825 |
0.27 |
| chr3_161194366_161194517 | 0.21 |
OTOL1 |
otolin 1 |
20155 |
0.28 |
| chr6_106534568_106534850 | 0.21 |
PRDM1 |
PR domain containing 1, with ZNF domain |
514 |
0.83 |
| chr1_96441392_96441543 | 0.21 |
RP11-147C23.1 |
Uncharacterized protein |
16078 |
0.28 |
| chr2_64320008_64320159 | 0.21 |
AC074289.1 |
|
50290 |
0.14 |
| chr2_228712880_228713031 | 0.21 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
22815 |
0.18 |
| chr20_48419063_48419214 | 0.21 |
ENSG00000252540 |
. |
7790 |
0.16 |
| chr8_29959377_29959528 | 0.21 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
4 |
0.97 |
| chr13_42970913_42971064 | 0.21 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
124699 |
0.06 |
| chr14_21099530_21099742 | 0.21 |
OR6S1 |
olfactory receptor, family 6, subfamily S, member 1 |
10214 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.3 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.5 | GO:0002363 | alpha-beta T cell lineage commitment(GO:0002363) |
| 0.1 | 0.3 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.5 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.1 | 0.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.5 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.5 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.0 | GO:0051571 | positive regulation of histone methylation(GO:0031062) positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0048670 | collateral sprouting(GO:0048668) regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.2 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.3 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0071028 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.3 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.7 | GO:0072401 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.6 | GO:0097194 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.0 | GO:0033080 | immature T cell proliferation in thymus(GO:0033080) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:1902750 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0071331 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.3 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.0 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0051380 | beta-adrenergic receptor activity(GO:0004939) norepinephrine binding(GO:0051380) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |