Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
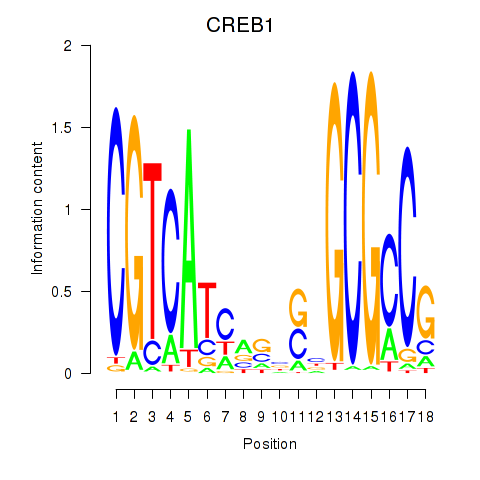
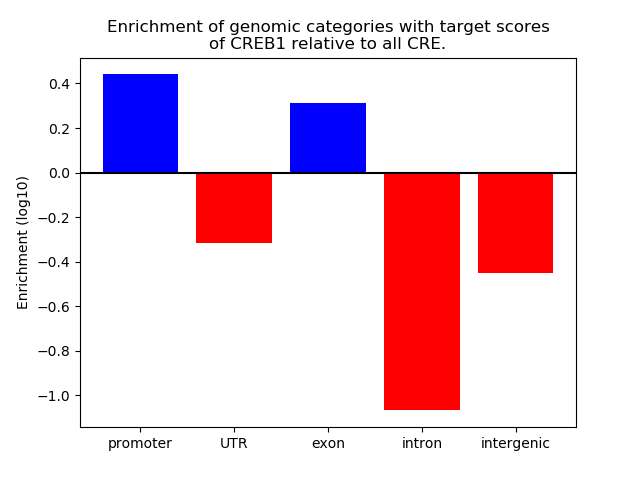
Results for CREB1
Z-value: 0.33
Transcription factors associated with CREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB1
|
ENSG00000118260.10 | cAMP responsive element binding protein 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
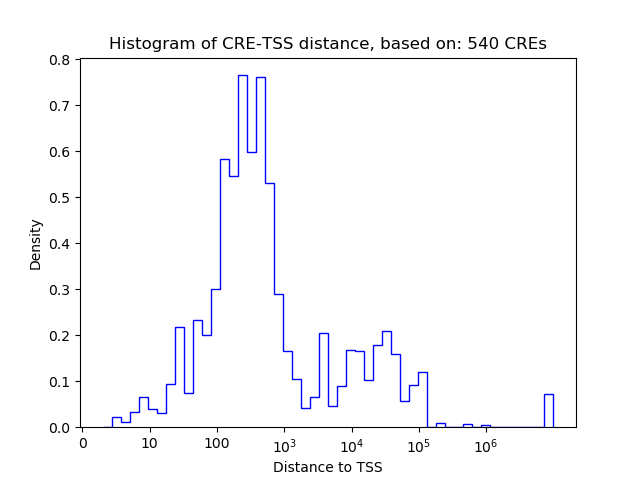
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_208365436_208365587 | CREB1 | 28950 | 0.179353 | 0.74 | 2.1e-02 | Click! |
| chr2_208395473_208395722 | CREB1 | 769 | 0.520090 | 0.60 | 8.9e-02 | Click! |
| chr2_208397856_208398007 | CREB1 | 3103 | 0.254747 | -0.60 | 9.0e-02 | Click! |
| chr2_208462214_208462365 | CREB1 | 22279 | 0.163369 | 0.59 | 9.2e-02 | Click! |
| chr2_208395832_208396657 | CREB1 | 1416 | 0.373902 | 0.57 | 1.1e-01 | Click! |
Activity of the CREB1 motif across conditions
Conditions sorted by the z-value of the CREB1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
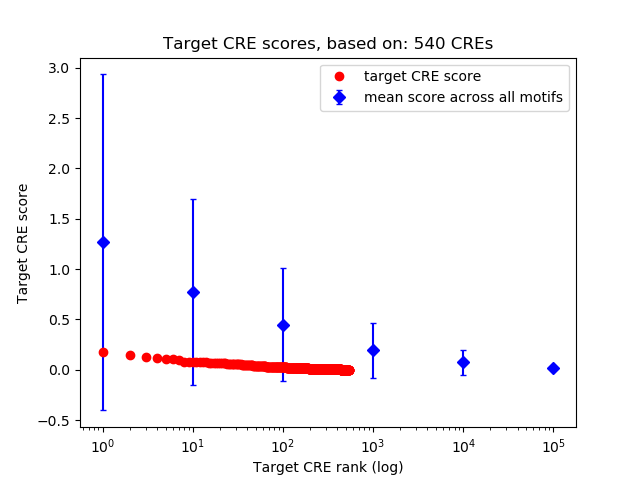
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_20491916_20492779 | 0.18 |
CDRT15L2 |
CMT1A duplicated region transcript 15-like 2 |
9310 |
0.21 |
| chr17_15689649_15690074 | 0.15 |
ENSG00000251829 |
. |
4206 |
0.19 |
| chr13_109792800_109793227 | 0.13 |
MYO16-AS2 |
MYO16 antisense RNA 2 |
38746 |
0.2 |
| chr18_74773875_74774026 | 0.11 |
MBP |
myelin basic protein |
43267 |
0.18 |
| chr1_57044587_57045121 | 0.11 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
387 |
0.91 |
| chr21_46961161_46962032 | 0.10 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
156 |
0.96 |
| chr6_116692035_116692441 | 0.10 |
DSE |
dermatan sulfate epimerase |
128 |
0.97 |
| chr11_49455905_49456091 | 0.08 |
RP11-707M1.1 |
|
126372 |
0.06 |
| chr4_39367062_39368041 | 0.08 |
RFC1 |
replication factor C (activator 1) 1, 145kDa |
418 |
0.81 |
| chrX_20135177_20135375 | 0.08 |
MAP7D2 |
MAP7 domain containing 2 |
241 |
0.9 |
| chr8_687260_687856 | 0.08 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
93 |
0.97 |
| chr2_190444828_190446170 | 0.08 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
114 |
0.98 |
| chr9_17578628_17579355 | 0.08 |
SH3GL2 |
SH3-domain GRB2-like 2 |
130 |
0.98 |
| chr20_32273512_32274295 | 0.07 |
E2F1 |
E2F transcription factor 1 |
307 |
0.81 |
| chr19_41931658_41931809 | 0.07 |
CTC-435M10.6 |
|
469 |
0.62 |
| chr14_92789494_92790350 | 0.07 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
27 |
0.99 |
| chr5_142782115_142782536 | 0.07 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
23 |
0.99 |
| chr3_154145836_154146602 | 0.07 |
GPR149 |
G protein-coupled receptor 149 |
1285 |
0.6 |
| chrX_334679_335404 | 0.07 |
PPP2R3B |
protein phosphatase 2, regulatory subunit B'', beta |
262 |
0.94 |
| chr12_93323322_93324013 | 0.06 |
EEA1 |
early endosome antigen 1 |
560 |
0.8 |
| chr11_118230043_118230305 | 0.06 |
UBE4A |
ubiquitination factor E4A |
126 |
0.93 |
| chr3_53276217_53276368 | 0.06 |
TKT |
transketolase |
13724 |
0.14 |
| chr3_112279714_112280459 | 0.06 |
SLC35A5 |
solute carrier family 35, member A5 |
470 |
0.59 |
| chrY_284683_285373 | 0.06 |
NA |
NA |
> 106 |
NA |
| chr7_37487305_37488000 | 0.06 |
ELMO1 |
engulfment and cell motility 1 |
901 |
0.61 |
| chr19_10527476_10527700 | 0.06 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
31 |
0.95 |
| chr21_35987336_35987662 | 0.06 |
RCAN1 |
regulator of calcineurin 1 |
58 |
0.98 |
| chr15_74657875_74658107 | 0.06 |
CTD-2311M21.3 |
|
215 |
0.75 |
| chr2_60962956_60963147 | 0.06 |
ENSG00000252718 |
. |
16276 |
0.19 |
| chr17_17380267_17380681 | 0.06 |
MED9 |
mediator complex subunit 9 |
166 |
0.94 |
| chr7_119450_119601 | 0.05 |
FAM20C |
family with sequence similarity 20, member C |
73444 |
0.11 |
| chr2_130939128_130939480 | 0.05 |
MZT2B |
mitotic spindle organizing protein 2B |
6 |
0.62 |
| chr18_53446598_53447540 | 0.05 |
TCF4 |
transcription factor 4 |
115051 |
0.07 |
| chr7_23145115_23145340 | 0.05 |
KLHL7 |
kelch-like family member 7 |
126 |
0.97 |
| chrX_132548696_132549413 | 0.05 |
GPC4 |
glypican 4 |
464 |
0.88 |
| chr3_71834358_71834666 | 0.05 |
PROK2 |
prokineticin 2 |
155 |
0.96 |
| chr15_44092835_44093206 | 0.05 |
HYPK |
huntingtin interacting protein K |
223 |
0.57 |
| chr17_34891354_34891543 | 0.05 |
PIGW |
phosphatidylinositol glycan anchor biosynthesis, class W |
45 |
0.91 |
| chr1_9599629_9600173 | 0.05 |
SLC25A33 |
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
360 |
0.89 |
| chr8_59323967_59324678 | 0.05 |
UBXN2B |
UBX domain protein 2B |
499 |
0.88 |
| chr17_1532581_1533057 | 0.05 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
689 |
0.52 |
| chr12_133022737_133022888 | 0.05 |
MUC8 |
mucin 8 |
27914 |
0.17 |
| chr16_87526385_87526690 | 0.05 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
886 |
0.63 |
| chr20_47443528_47443935 | 0.05 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
689 |
0.79 |
| chr1_109940210_109941121 | 0.04 |
SORT1 |
sortilin 1 |
92 |
0.96 |
| chr1_61549203_61549354 | 0.04 |
ENSG00000263380 |
. |
76 |
0.94 |
| chr2_26785075_26785548 | 0.04 |
C2orf70 |
chromosome 2 open reading frame 70 |
139 |
0.96 |
| chr2_64836667_64836835 | 0.04 |
ENSG00000252414 |
. |
30725 |
0.17 |
| chr4_134072879_134073697 | 0.04 |
PCDH10 |
protocadherin 10 |
2818 |
0.44 |
| chr9_34049001_34049202 | 0.04 |
UBAP2 |
ubiquitin associated protein 2 |
154 |
0.89 |
| chr7_4901206_4901513 | 0.04 |
PAPOLB |
poly(A) polymerase beta (testis specific) |
266 |
0.91 |
| chr11_9595385_9595551 | 0.04 |
WEE1 |
WEE1 G2 checkpoint kinase |
240 |
0.9 |
| chr7_140623410_140624534 | 0.04 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
592 |
0.79 |
| chrX_39548256_39548819 | 0.04 |
ENSG00000263730 |
. |
28067 |
0.24 |
| chr7_39894513_39894809 | 0.04 |
ENSG00000242855 |
. |
33311 |
0.18 |
| chr1_12040274_12040953 | 0.04 |
MFN2 |
mitofusin 2 |
244 |
0.89 |
| chr9_96339036_96339199 | 0.04 |
PHF2 |
PHD finger protein 2 |
208 |
0.94 |
| chr3_195384928_195385957 | 0.04 |
SDHAP2 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2 |
475 |
0.81 |
| chrX_71933533_71934701 | 0.04 |
PHKA1 |
phosphorylase kinase, alpha 1 (muscle) |
11 |
0.97 |
| chr16_89159943_89160452 | 0.03 |
ACSF3 |
acyl-CoA synthetase family member 3 |
20 |
0.97 |
| chr3_143689644_143690155 | 0.03 |
C3orf58 |
chromosome 3 open reading frame 58 |
741 |
0.8 |
| chr3_87842154_87842380 | 0.03 |
ENSG00000200703 |
. |
50468 |
0.17 |
| chr2_27008928_27009181 | 0.03 |
CENPA |
centromere protein A |
118 |
0.96 |
| chr5_170289344_170289797 | 0.03 |
RANBP17 |
RAN binding protein 17 |
696 |
0.71 |
| chr18_52988378_52989650 | 0.03 |
TCF4 |
transcription factor 4 |
203 |
0.97 |
| chr11_74442538_74442948 | 0.03 |
CHRDL2 |
chordin-like 2 |
313 |
0.84 |
| chr3_49055259_49056395 | 0.03 |
DALRD3 |
DALR anticodon binding domain containing 3 |
170 |
0.84 |
| chr2_74691535_74692741 | 0.03 |
MOGS |
mannosyl-oligosaccharide glucosidase |
351 |
0.67 |
| chrX_138285953_138286466 | 0.03 |
FGF13 |
fibroblast growth factor 13 |
60 |
0.99 |
| chr17_72732981_72733167 | 0.03 |
RAB37 |
RAB37, member RAS oncogene family |
18 |
0.95 |
| chr5_76506770_76507546 | 0.03 |
PDE8B |
phosphodiesterase 8B |
407 |
0.88 |
| chr1_109642808_109643473 | 0.03 |
ENSG00000270066 |
. |
325 |
0.81 |
| chr4_6783649_6783989 | 0.03 |
KIAA0232 |
KIAA0232 |
550 |
0.79 |
| chr15_70391403_70391876 | 0.03 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
1124 |
0.58 |
| chr4_174459155_174459419 | 0.03 |
HAND2-AS1 |
HAND2 antisense RNA 1 (head to head) |
6591 |
0.22 |
| chr15_80696207_80696642 | 0.03 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
269 |
0.71 |
| chr11_65547664_65548343 | 0.03 |
AP5B1 |
adaptor-related protein complex 5, beta 1 subunit |
270 |
0.82 |
| chr16_52580478_52580769 | 0.03 |
TOX3 |
TOX high mobility group box family member 3 |
297 |
0.95 |
| chr3_69591903_69592054 | 0.03 |
FRMD4B |
FERM domain containing 4B |
244 |
0.96 |
| chr18_74534859_74535018 | 0.03 |
ZNF236 |
zinc finger protein 236 |
375 |
0.68 |
| chr1_245134150_245135065 | 0.03 |
RP11-156E8.1 |
Uncharacterized protein |
217 |
0.85 |
| chr3_125899243_125899606 | 0.03 |
ALDH1L1 |
aldehyde dehydrogenase 1 family, member L1 |
5 |
0.9 |
| chr7_3340905_3341061 | 0.03 |
SDK1 |
sidekick cell adhesion molecule 1 |
97 |
0.95 |
| chr2_100105997_100106148 | 0.03 |
REV1 |
REV1, polymerase (DNA directed) |
363 |
0.89 |
| chr10_130008645_130009418 | 0.03 |
MKI67 |
marker of proliferation Ki-67 |
84382 |
0.11 |
| chr1_54410859_54411299 | 0.03 |
HSPB11 |
heat shock protein family B (small), member 11 |
166 |
0.86 |
| chr9_131464565_131465241 | 0.03 |
PKN3 |
protein kinase N3 |
101 |
0.93 |
| chr13_110437375_110437816 | 0.03 |
IRS2 |
insulin receptor substrate 2 |
1320 |
0.57 |
| chr16_1359047_1359364 | 0.03 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
51 |
0.94 |
| chr14_50235014_50236034 | 0.03 |
KLHDC2 |
kelch domain containing 2 |
638 |
0.62 |
| chr10_104210416_104210610 | 0.03 |
ENSG00000269609 |
. |
125 |
0.87 |
| chr2_7057645_7057806 | 0.03 |
RNF144A |
ring finger protein 144A |
202 |
0.9 |
| chr9_140500205_140501463 | 0.03 |
ARRDC1 |
arrestin domain containing 1 |
679 |
0.56 |
| chr17_74534170_74534513 | 0.03 |
CYGB |
cytoglobin |
354 |
0.7 |
| chr2_136289143_136290038 | 0.03 |
R3HDM1 |
R3H domain containing 1 |
503 |
0.62 |
| chr16_1470975_1471179 | 0.03 |
C16orf91 |
chromosome 16 open reading frame 91 |
276 |
0.79 |
| chr3_69591491_69591662 | 0.02 |
FRMD4B |
FERM domain containing 4B |
158 |
0.98 |
| chr2_190305327_190305991 | 0.02 |
WDR75 |
WD repeat domain 75 |
500 |
0.87 |
| chr12_69633375_69633694 | 0.02 |
CPSF6 |
cleavage and polyadenylation specific factor 6, 68kDa |
112 |
0.98 |
| chr2_97482659_97482901 | 0.02 |
CNNM3 |
cyclin M3 |
793 |
0.53 |
| chr16_3108546_3109349 | 0.02 |
RP11-473M20.7 |
|
139 |
0.86 |
| chr10_103577050_103577916 | 0.02 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
140 |
0.93 |
| chr6_56820015_56820822 | 0.02 |
BEND6 |
BEN domain containing 6 |
248 |
0.86 |
| chr19_8008965_8009156 | 0.02 |
TIMM44 |
translocase of inner mitochondrial membrane 44 homolog (yeast) |
255 |
0.8 |
| chr11_63766571_63766823 | 0.02 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
12383 |
0.11 |
| chr8_21946490_21946833 | 0.02 |
FAM160B2 |
family with sequence similarity 160, member B2 |
58 |
0.96 |
| chr16_54971520_54971850 | 0.02 |
IRX5 |
iroquois homeobox 5 |
5701 |
0.35 |
| chr2_85938999_85939212 | 0.02 |
ENSG00000199687 |
. |
16355 |
0.12 |
| chr22_20104378_20104805 | 0.02 |
TRMT2A |
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
154 |
0.7 |
| chr12_115889903_115890336 | 0.02 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
89302 |
0.1 |
| chr15_52860024_52861356 | 0.02 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
339 |
0.87 |
| chr1_63782781_63783608 | 0.02 |
RP4-792G4.2 |
|
4006 |
0.2 |
| chr1_1589957_1590616 | 0.02 |
CDK11B |
cyclin-dependent kinase 11B |
187 |
0.75 |
| chr1_22108808_22109469 | 0.02 |
USP48 |
ubiquitin specific peptidase 48 |
346 |
0.89 |
| chr7_56019535_56020015 | 0.02 |
MRPS17 |
mitochondrial ribosomal protein S17 |
119 |
0.72 |
| chr11_568685_568932 | 0.02 |
ENSG00000199038 |
. |
610 |
0.48 |
| chr19_13264252_13264621 | 0.02 |
CTC-250I14.6 |
|
90 |
0.93 |
| chr9_132382165_132382732 | 0.02 |
C9orf50 |
chromosome 9 open reading frame 50 |
607 |
0.62 |
| chr7_99156308_99157048 | 0.02 |
ZNF655 |
zinc finger protein 655 |
193 |
0.77 |
| chr20_48595704_48595927 | 0.02 |
SNAI1 |
snail family zinc finger 1 |
3721 |
0.18 |
| chr7_113724609_113725113 | 0.02 |
FOXP2 |
forkhead box P2 |
1754 |
0.44 |
| chr10_108924530_108924681 | 0.02 |
SORCS1 |
sortilin-related VPS10 domain containing receptor 1 |
313 |
0.95 |
| chr11_77185235_77185792 | 0.02 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
167 |
0.92 |
| chr2_61696387_61697458 | 0.02 |
USP34 |
ubiquitin specific peptidase 34 |
982 |
0.45 |
| chr20_3994903_3996170 | 0.02 |
RNF24 |
ring finger protein 24 |
500 |
0.83 |
| chr1_208132304_208132634 | 0.02 |
CD34 |
CD34 molecule |
47722 |
0.19 |
| chr15_74219902_74220340 | 0.02 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
6 |
0.96 |
| chr16_3207596_3208760 | 0.02 |
CASP16 |
caspase 16, apoptosis-related cysteine peptidase (putative) |
13934 |
0.08 |
| chr2_176944534_176944961 | 0.02 |
EVX2 |
even-skipped homeobox 2 |
3894 |
0.12 |
| chr12_104531819_104531970 | 0.02 |
NFYB |
nuclear transcription factor Y, beta |
87 |
0.97 |
| chr1_90460754_90461861 | 0.02 |
ZNF326 |
zinc finger protein 326 |
580 |
0.8 |
| chr10_112403975_112404246 | 0.02 |
RBM20 |
RNA binding motif protein 20 |
45 |
0.96 |
| chr1_233463556_233464041 | 0.02 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
284 |
0.94 |
| chrX_119149654_119149897 | 0.02 |
RP4-755D9.1 |
|
20426 |
0.16 |
| chr9_134954308_134955306 | 0.02 |
MED27 |
mediator complex subunit 27 |
446 |
0.89 |
| chr9_130699622_130700109 | 0.02 |
DPM2 |
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
257 |
0.5 |
| chr4_74904304_74905084 | 0.02 |
CXCL3 |
chemokine (C-X-C motif) ligand 3 |
288 |
0.87 |
| chrY_21238831_21239141 | 0.02 |
ENSG00000252766 |
. |
58013 |
0.16 |
| chr4_96469881_96470326 | 0.02 |
UNC5C |
unc-5 homolog C (C. elegans) |
20 |
0.69 |
| chr3_142607235_142608274 | 0.02 |
PCOLCE2 |
procollagen C-endopeptidase enhancer 2 |
13 |
0.98 |
| chr14_33408703_33409010 | 0.02 |
NPAS3 |
neuronal PAS domain protein 3 |
333 |
0.94 |
| chr8_10696975_10697533 | 0.02 |
PINX1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
27 |
0.57 |
| chr1_9882638_9884014 | 0.02 |
CLSTN1 |
calsyntenin 1 |
716 |
0.65 |
| chr8_135725075_135725586 | 0.02 |
ZFAT |
zinc finger and AT hook domain containing |
49 |
0.99 |
| chr19_56156604_56156918 | 0.02 |
ZNF581 |
zinc finger protein 581 |
1585 |
0.15 |
| chr9_77642770_77643186 | 0.02 |
C9orf41 |
chromosome 9 open reading frame 41 |
211 |
0.93 |
| chr11_8290078_8290567 | 0.02 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
59 |
0.98 |
| chr15_68155525_68155920 | 0.02 |
ENSG00000206625 |
. |
23339 |
0.22 |
| chr2_223155673_223155824 | 0.02 |
CCDC140 |
coiled-coil domain containing 140 |
7118 |
0.2 |
| chr2_27578903_27579940 | 0.02 |
GTF3C2 |
general transcription factor IIIC, polypeptide 2, beta 110kDa |
62 |
0.77 |
| chr2_152493968_152494122 | 0.02 |
NEB |
nebulin |
53912 |
0.17 |
| chr16_88870149_88870541 | 0.02 |
CDT1 |
chromatin licensing and DNA replication factor 1 |
724 |
0.48 |
| chr7_155249437_155250320 | 0.02 |
EN2 |
engrailed homeobox 2 |
946 |
0.59 |
| chr6_170921254_170921775 | 0.02 |
PDCD2 |
programmed cell death 2 |
27766 |
0.19 |
| chr2_238395971_238396408 | 0.02 |
MLPH |
melanophilin |
270 |
0.92 |
| chr13_53024209_53024738 | 0.02 |
VPS36 |
vacuolar protein sorting 36 homolog (S. cerevisiae) |
284 |
0.9 |
| chr10_28287400_28288216 | 0.02 |
ARMC4 |
armadillo repeat containing 4 |
169 |
0.97 |
| chr3_100053843_100054079 | 0.02 |
NIT2 |
nitrilase family, member 2 |
306 |
0.91 |
| chrX_334213_334384 | 0.02 |
PPP2R3B |
protein phosphatase 2, regulatory subunit B'', beta |
481 |
0.86 |
| chr15_42840348_42841494 | 0.02 |
LRRC57 |
leucine rich repeat containing 57 |
49 |
0.54 |
| chr14_100706520_100706671 | 0.02 |
YY1 |
YY1 transcription factor |
499 |
0.64 |
| chr6_101849901_101850127 | 0.02 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
2860 |
0.43 |
| chr12_51818617_51818968 | 0.02 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
109 |
0.81 |
| chrY_284220_284379 | 0.02 |
NA |
NA |
> 106 |
NA |
| chr1_41249080_41249567 | 0.02 |
KCNQ4 |
potassium voltage-gated channel, KQT-like subfamily, member 4 |
361 |
0.82 |
| chr3_197354198_197354750 | 0.02 |
AC024560.3 |
|
245 |
0.93 |
| chr20_33413142_33413293 | 0.02 |
NCOA6 |
nuclear receptor coactivator 6 |
235 |
0.93 |
| chrX_153402310_153402730 | 0.01 |
OPN1LW |
opsin 1 (cone pigments), long-wave-sensitive |
7178 |
0.14 |
| chr22_51020587_51021277 | 0.01 |
CHKB |
choline kinase beta |
496 |
0.39 |
| chr8_41997342_41997944 | 0.01 |
RP11-589C21.5 |
|
12638 |
0.17 |
| chr6_137244870_137245021 | 0.01 |
SLC35D3 |
solute carrier family 35, member D3 |
1543 |
0.4 |
| chr2_11051733_11051998 | 0.01 |
KCNF1 |
potassium voltage-gated channel, subfamily F, member 1 |
198 |
0.95 |
| chr5_141404364_141404742 | 0.01 |
GNPDA1 |
glucosamine-6-phosphate deaminase 1 |
11947 |
0.17 |
| chr20_307052_307593 | 0.01 |
SOX12 |
SRY (sex determining region Y)-box 12 |
1083 |
0.33 |
| chr11_73498863_73499083 | 0.01 |
MRPL48 |
mitochondrial ribosomal protein L48 |
2 |
0.97 |
| chr12_95942120_95942537 | 0.01 |
USP44 |
ubiquitin specific peptidase 44 |
246 |
0.94 |
| chr11_64569817_64570601 | 0.01 |
MAP4K2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
438 |
0.72 |
| chr11_123525466_123525691 | 0.01 |
SCN3B |
sodium channel, voltage-gated, type III, beta subunit |
136 |
0.96 |
| chr12_112546417_112546609 | 0.01 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
34 |
0.97 |
| chr17_7144804_7145679 | 0.01 |
GABARAP |
GABA(A) receptor-associated protein |
103 |
0.89 |
| chr17_8042262_8042413 | 0.01 |
PER1 |
period circadian clock 1 |
7005 |
0.08 |
| chr10_70588000_70588364 | 0.01 |
STOX1 |
storkhead box 1 |
619 |
0.68 |
| chr17_37910669_37910916 | 0.01 |
GRB7 |
growth factor receptor-bound protein 7 |
12287 |
0.11 |
| chr7_35076877_35077068 | 0.01 |
DPY19L1 |
dpy-19-like 1 (C. elegans) |
681 |
0.8 |
| chr2_71115848_71116159 | 0.01 |
VAX2 |
ventral anterior homeobox 2 |
11717 |
0.13 |
| chr3_96531977_96533205 | 0.01 |
EPHA6 |
EPH receptor A6 |
834 |
0.76 |
| chr11_2466604_2466835 | 0.01 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
498 |
0.74 |
| chr16_84149538_84150449 | 0.01 |
MBTPS1 |
membrane-bound transcription factor peptidase, site 1 |
407 |
0.6 |
| chr16_46660648_46660928 | 0.01 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
5250 |
0.2 |
| chr12_133405892_133406043 | 0.01 |
GOLGA3 |
golgin A3 |
528 |
0.72 |
| chr10_12392249_12392465 | 0.01 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
625 |
0.72 |
| chr2_28616306_28616795 | 0.01 |
FOSL2 |
FOS-like antigen 2 |
825 |
0.43 |
| chr15_99557942_99558631 | 0.01 |
PGPEP1L |
pyroglutamyl-peptidase I-like |
7262 |
0.22 |
| chr10_32636312_32636863 | 0.01 |
RP11-135A24.2 |
|
262 |
0.56 |
| chr1_17248321_17248578 | 0.01 |
CROCC |
ciliary rootlet coiled-coil, rootletin |
4 |
0.97 |
| chr16_66785084_66785300 | 0.01 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
305 |
0.86 |
| chr17_18086808_18086984 | 0.01 |
ALKBH5 |
alkB, alkylation repair homolog 5 (E. coli) |
504 |
0.6 |
| chr2_92027935_92028159 | 0.01 |
IGKV1OR-1 |
immunoglobulin kappa variable 1/OR-1 (pseudogene) |
22065 |
0.23 |
| chr19_47951006_47951312 | 0.01 |
SLC8A2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
23961 |
0.12 |
| chr1_64059512_64060140 | 0.01 |
PGM1 |
phosphoglucomutase 1 |
346 |
0.87 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |