Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
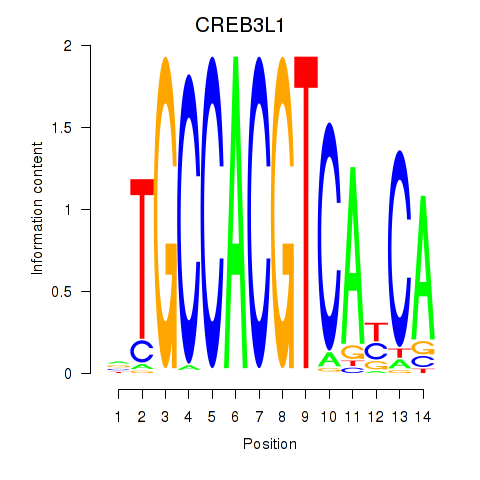
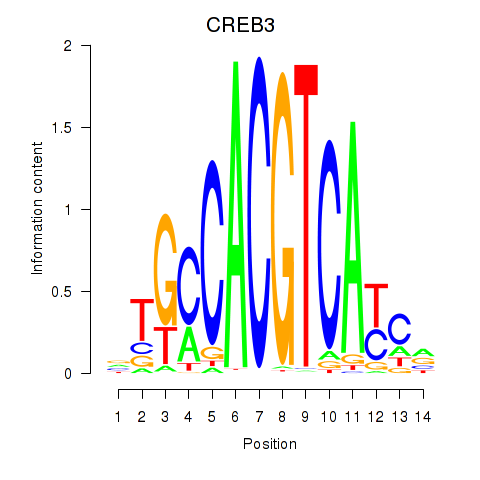
Results for CREB3L1_CREB3
Z-value: 0.82
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
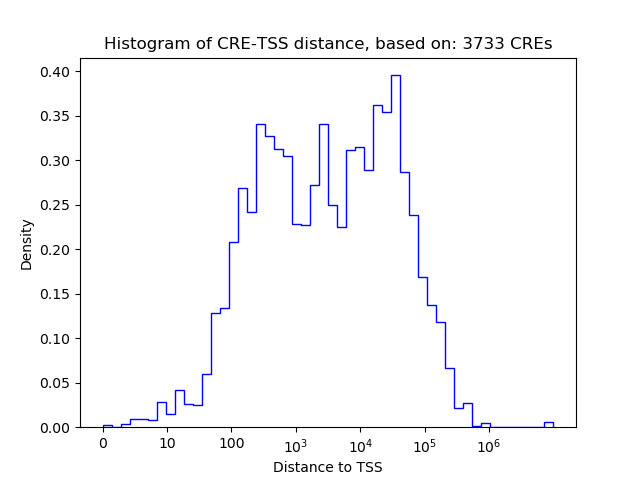
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_35728074_35728276 | CREB3 | 4157 | 0.088840 | 0.61 | 8.2e-02 | Click! |
| chr9_35727770_35727988 | CREB3 | 4453 | 0.086846 | 0.56 | 1.2e-01 | Click! |
| chr9_35727615_35727766 | CREB3 | 4642 | 0.085752 | 0.55 | 1.2e-01 | Click! |
| chr9_35727078_35727229 | CREB3 | 5179 | 0.083219 | 0.50 | 1.7e-01 | Click! |
| chr9_35714007_35714158 | CREB3 | 18250 | 0.066425 | -0.46 | 2.1e-01 | Click! |
| chr11_46307086_46307288 | CREB3L1 | 7959 | 0.176890 | 0.51 | 1.6e-01 | Click! |
| chr11_46300696_46300847 | CREB3L1 | 1543 | 0.366749 | 0.50 | 1.7e-01 | Click! |
| chr11_46300240_46300518 | CREB3L1 | 1151 | 0.464926 | 0.42 | 2.6e-01 | Click! |
| chr11_46340567_46340770 | CREB3L1 | 7960 | 0.160269 | -0.42 | 2.6e-01 | Click! |
| chr11_46302791_46303236 | CREB3L1 | 3785 | 0.211708 | 0.38 | 3.2e-01 | Click! |
Activity of the CREB3L1_CREB3 motif across conditions
Conditions sorted by the z-value of the CREB3L1_CREB3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
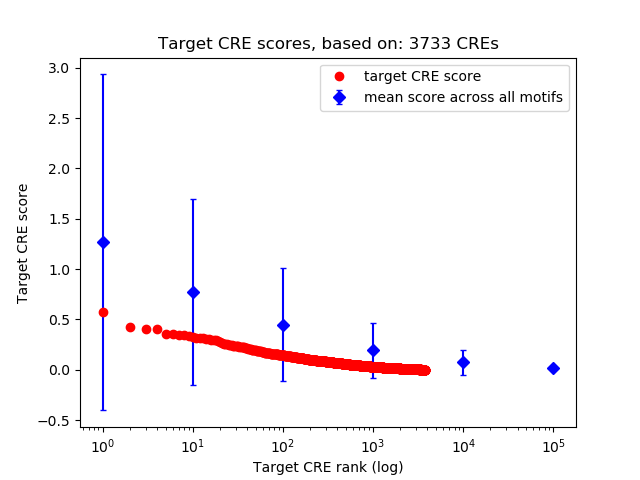
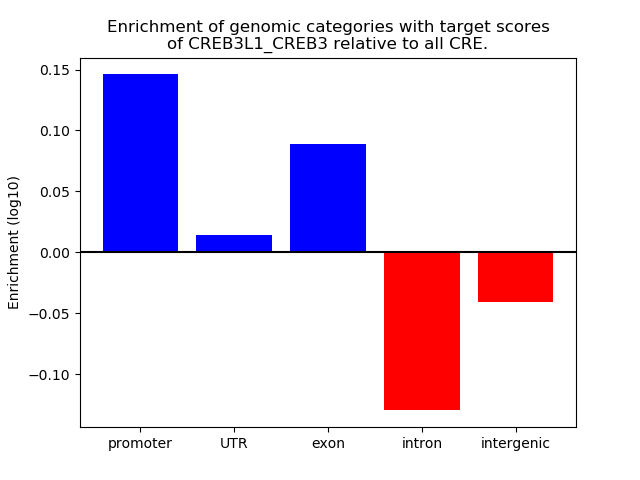
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_15722330_15722494 | 0.58 |
MEOX2 |
mesenchyme homeobox 2 |
4025 |
0.29 |
| chr16_1014713_1014988 | 0.43 |
LMF1 |
lipase maturation factor 1 |
728 |
0.55 |
| chr1_170632304_170632495 | 0.41 |
PRRX1 |
paired related homeobox 1 |
124 |
0.98 |
| chr8_70042166_70042317 | 0.40 |
ENSG00000238808 |
. |
19432 |
0.3 |
| chr7_100200956_100201118 | 0.36 |
PCOLCE-AS1 |
PCOLCE antisense RNA 1 |
379 |
0.65 |
| chr5_141443601_141443885 | 0.36 |
NDFIP1 |
Nedd4 family interacting protein 1 |
44327 |
0.13 |
| chr1_27445743_27446018 | 0.34 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
35093 |
0.15 |
| chr15_99259160_99259373 | 0.34 |
IGF1R |
insulin-like growth factor 1 receptor |
8206 |
0.26 |
| chr2_62444590_62444741 | 0.33 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
2012 |
0.3 |
| chr9_80055112_80055292 | 0.33 |
RP11-466A17.1 |
|
16009 |
0.24 |
| chr4_152146952_152147499 | 0.32 |
SH3D19 |
SH3 domain containing 19 |
7 |
0.98 |
| chr10_6125711_6125919 | 0.32 |
RBM17 |
RNA binding motif protein 17 |
5135 |
0.17 |
| chr8_66953995_66954146 | 0.31 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
1642 |
0.5 |
| chr2_198540490_198540682 | 0.31 |
RFTN2 |
raftlin family member 2 |
133 |
0.96 |
| chr9_137288070_137288221 | 0.31 |
RXRA |
retinoid X receptor, alpha |
10283 |
0.23 |
| chr5_171560548_171560699 | 0.30 |
ENSG00000266671 |
. |
52643 |
0.12 |
| chr12_129291969_129292120 | 0.29 |
SLC15A4 |
solute carrier family 15 (oligopeptide transporter), member 4 |
2859 |
0.35 |
| chr16_88658218_88658369 | 0.29 |
ZC3H18 |
zinc finger CCCH-type containing 18 |
5268 |
0.15 |
| chr1_204010490_204010911 | 0.29 |
SOX13 |
SRY (sex determining region Y)-box 13 |
32032 |
0.15 |
| chr2_218221473_218221860 | 0.27 |
ENSG00000251982 |
. |
105387 |
0.08 |
| chr2_218867466_218867778 | 0.27 |
TNS1 |
tensin 1 |
96 |
0.97 |
| chr20_44463514_44463665 | 0.26 |
SNX21 |
sorting nexin family member 21 |
814 |
0.34 |
| chr1_23883319_23883470 | 0.26 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
2891 |
0.25 |
| chr17_57407133_57407390 | 0.26 |
YPEL2 |
yippee-like 2 (Drosophila) |
1802 |
0.33 |
| chr3_27523238_27523389 | 0.25 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
2525 |
0.3 |
| chr7_23274495_23274692 | 0.24 |
GPNMB |
glycoprotein (transmembrane) nmb |
11589 |
0.2 |
| chr1_12235343_12235565 | 0.24 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
8394 |
0.16 |
| chr21_17445369_17445660 | 0.24 |
ENSG00000252273 |
. |
37685 |
0.23 |
| chr10_44879097_44879375 | 0.24 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
1255 |
0.52 |
| chr2_101541390_101541618 | 0.23 |
NPAS2 |
neuronal PAS domain protein 2 |
104 |
0.97 |
| chr1_224008752_224008903 | 0.23 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
6797 |
0.28 |
| chr16_89830973_89831469 | 0.23 |
FANCA |
Fanconi anemia, complementation group A |
19751 |
0.11 |
| chrX_23686647_23686860 | 0.23 |
PRDX4 |
peroxiredoxin 4 |
1085 |
0.58 |
| chr1_66846122_66846273 | 0.23 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
26132 |
0.25 |
| chr11_63686882_63687128 | 0.22 |
RCOR2 |
REST corepressor 2 |
2689 |
0.17 |
| chr6_133254252_133254403 | 0.22 |
ENSG00000200534 |
. |
115969 |
0.05 |
| chr6_117717394_117717545 | 0.22 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
29549 |
0.15 |
| chrX_100872700_100873033 | 0.22 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
125 |
0.93 |
| chr10_124254663_124255115 | 0.22 |
HTRA1 |
HtrA serine peptidase 1 |
11318 |
0.2 |
| chr17_66082289_66082451 | 0.22 |
LINC00674 |
long intergenic non-protein coding RNA 674 |
15679 |
0.18 |
| chr1_178991033_178991223 | 0.21 |
FAM20B |
family with sequence similarity 20, member B |
3811 |
0.29 |
| chr1_170640082_170640341 | 0.21 |
PRRX1 |
paired related homeobox 1 |
7133 |
0.3 |
| chr1_90331815_90331966 | 0.20 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
19751 |
0.22 |
| chr19_42130849_42131075 | 0.20 |
CEACAM4 |
carcinoembryonic antigen-related cell adhesion molecule 4 |
2480 |
0.23 |
| chr5_121509459_121509610 | 0.20 |
CTC-441N14.1 |
|
17824 |
0.18 |
| chr6_148830339_148830689 | 0.20 |
ENSG00000223322 |
. |
14862 |
0.29 |
| chr20_19956104_19956326 | 0.19 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
41545 |
0.15 |
| chr2_208199066_208199401 | 0.19 |
ENSG00000221628 |
. |
65085 |
0.12 |
| chr6_27573492_27573857 | 0.19 |
ENSG00000206671 |
. |
9483 |
0.2 |
| chr4_11652300_11652527 | 0.19 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
221024 |
0.02 |
| chr6_1885704_1885932 | 0.19 |
FOXC1 |
forkhead box C1 |
275137 |
0.01 |
| chr17_43312812_43312963 | 0.19 |
FMNL1 |
formin-like 1 |
5555 |
0.1 |
| chr3_72940504_72940835 | 0.19 |
GXYLT2 |
glucoside xylosyltransferase 2 |
3445 |
0.28 |
| chr1_217262846_217263329 | 0.19 |
ESRRG |
estrogen-related receptor gamma |
111 |
0.98 |
| chr5_88171366_88171720 | 0.19 |
MEF2C |
myocyte enhancer factor 2C |
2251 |
0.36 |
| chr8_23605481_23605654 | 0.19 |
ENSG00000216123 |
. |
20591 |
0.16 |
| chr3_45265689_45266072 | 0.19 |
TMEM158 |
transmembrane protein 158 (gene/pseudogene) |
1890 |
0.38 |
| chr10_45719589_45719755 | 0.18 |
ENSG00000207071 |
. |
35328 |
0.17 |
| chr5_1879697_1879902 | 0.18 |
IRX4 |
iroquois homeobox 4 |
3059 |
0.2 |
| chr4_114063101_114063433 | 0.18 |
ANK2 |
ankyrin 2, neuronal |
3521 |
0.33 |
| chr2_96658143_96658593 | 0.18 |
ANKRD36C |
ankyrin repeat domain 36C |
827 |
0.58 |
| chr5_92923285_92923507 | 0.17 |
ENSG00000237187 |
. |
2042 |
0.33 |
| chrX_41535546_41535697 | 0.17 |
ENSG00000242559 |
. |
7098 |
0.18 |
| chr5_151060620_151060937 | 0.17 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
4013 |
0.17 |
| chr12_52241613_52241902 | 0.17 |
AC068987.1 |
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
37968 |
0.13 |
| chr6_160876831_160877004 | 0.17 |
LPAL2 |
lipoprotein, Lp(a)-like 2, pseudogene |
55122 |
0.15 |
| chr8_9088633_9088784 | 0.17 |
ENSG00000207415 |
. |
56380 |
0.12 |
| chr17_6555060_6555332 | 0.17 |
C17orf100 |
chromosome 17 open reading frame 100 |
225 |
0.49 |
| chr5_172234489_172234691 | 0.16 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
26688 |
0.12 |
| chr17_66511128_66511617 | 0.16 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
108 |
0.97 |
| chr15_101728923_101729679 | 0.16 |
CHSY1 |
chondroitin sulfate synthase 1 |
62836 |
0.1 |
| chr2_85765868_85766150 | 0.16 |
MAT2A |
methionine adenosyltransferase II, alpha |
279 |
0.82 |
| chr3_194118884_194119077 | 0.16 |
GP5 |
glycoprotein V (platelet) |
103 |
0.96 |
| chr17_1959923_1960311 | 0.16 |
HIC1 |
hypermethylated in cancer 1 |
513 |
0.59 |
| chrX_128893788_128893939 | 0.16 |
SASH3 |
SAM and SH3 domain containing 3 |
20097 |
0.17 |
| chr18_22813939_22814258 | 0.16 |
ZNF521 |
zinc finger protein 521 |
2604 |
0.41 |
| chr17_49027559_49028056 | 0.16 |
SPAG9 |
sperm associated antigen 9 |
43491 |
0.13 |
| chr15_74988471_74988935 | 0.16 |
EDC3 |
enhancer of mRNA decapping 3 |
70 |
0.97 |
| chr2_3307191_3307353 | 0.16 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
2036 |
0.4 |
| chr3_72640881_72641032 | 0.16 |
ENSG00000199469 |
. |
15114 |
0.22 |
| chr20_36825206_36825357 | 0.16 |
TGM2 |
transglutaminase 2 |
30301 |
0.14 |
| chr6_149803641_149803913 | 0.15 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
492 |
0.77 |
| chr22_38864095_38864404 | 0.15 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
166 |
0.93 |
| chr20_56803890_56804242 | 0.15 |
ANKRD60 |
ankyrin repeat domain 60 |
357 |
0.88 |
| chr21_44061431_44061649 | 0.15 |
AP001626.2 |
|
9814 |
0.18 |
| chr1_204332360_204332939 | 0.15 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
3605 |
0.22 |
| chr2_110656312_110656910 | 0.15 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
343 |
0.93 |
| chr6_136547273_136547517 | 0.15 |
RP13-143G15.4 |
|
662 |
0.72 |
| chr19_39140182_39140489 | 0.15 |
ACTN4 |
actinin, alpha 4 |
1957 |
0.23 |
| chr5_54899055_54899206 | 0.15 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
68252 |
0.1 |
| chr16_31104780_31104975 | 0.15 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
55 |
0.91 |
| chr3_73673648_73674178 | 0.15 |
PDZRN3 |
PDZ domain containing ring finger 3 |
78 |
0.96 |
| chr4_177304632_177304846 | 0.15 |
RP11-87F15.2 |
|
63130 |
0.12 |
| chr20_24008090_24008393 | 0.15 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
38825 |
0.2 |
| chr16_86602293_86602507 | 0.15 |
RP11-463O9.5 |
|
1033 |
0.41 |
| chr7_37797263_37797414 | 0.15 |
GPR141 |
G protein-coupled receptor 141 |
17342 |
0.25 |
| chr7_37755543_37755694 | 0.15 |
GPR141 |
G protein-coupled receptor 141 |
24378 |
0.21 |
| chr2_109271526_109272143 | 0.15 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
325 |
0.91 |
| chr15_36699845_36700050 | 0.15 |
C15orf41 |
chromosome 15 open reading frame 41 |
171865 |
0.04 |
| chr4_155953158_155953309 | 0.15 |
RP11-92A5.2 |
|
176350 |
0.03 |
| chr20_47435468_47435675 | 0.15 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
8849 |
0.27 |
| chr6_13926565_13926716 | 0.14 |
RNF182 |
ring finger protein 182 |
1216 |
0.61 |
| chr8_29513803_29514023 | 0.14 |
ENSG00000221003 |
. |
272208 |
0.01 |
| chr20_61555717_61555868 | 0.14 |
DIDO1 |
death inducer-obliterator 1 |
2029 |
0.26 |
| chr15_68545515_68545666 | 0.14 |
CLN6 |
ceroid-lipofuscinosis, neuronal 6, late infantile, variant |
3854 |
0.2 |
| chr2_112250892_112251415 | 0.14 |
ENSG00000266139 |
. |
172485 |
0.03 |
| chrY_19868908_19869238 | 0.14 |
ENSG00000252513 |
. |
31969 |
0.22 |
| chr18_59537327_59537518 | 0.14 |
RNF152 |
ring finger protein 152 |
23570 |
0.27 |
| chr13_96297004_96297314 | 0.14 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
202 |
0.95 |
| chr7_38351039_38351457 | 0.14 |
STARD3NL |
STARD3 N-terminal like |
133251 |
0.05 |
| chr1_119521934_119522435 | 0.14 |
TBX15 |
T-box 15 |
8244 |
0.28 |
| chr3_150088154_150088521 | 0.14 |
TSC22D2 |
TSC22 domain family, member 2 |
37785 |
0.2 |
| chrY_2803195_2803357 | 0.14 |
ZFY |
zinc finger protein, Y-linked |
46 |
0.98 |
| chr2_89028504_89028655 | 0.14 |
RPIA |
ribose 5-phosphate isomerase A |
37417 |
0.1 |
| chr3_81766350_81766631 | 0.14 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
26290 |
0.28 |
| chr5_54898758_54898909 | 0.14 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
67955 |
0.1 |
| chr19_2775423_2775574 | 0.14 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
4719 |
0.12 |
| chr10_14592515_14592666 | 0.13 |
FAM107B |
family with sequence similarity 107, member B |
1921 |
0.42 |
| chr17_18061433_18061699 | 0.13 |
MYO15A |
myosin XVA |
56 |
0.96 |
| chr12_107350500_107350907 | 0.13 |
C12orf23 |
chromosome 12 open reading frame 23 |
140 |
0.94 |
| chr19_16438433_16439148 | 0.13 |
KLF2 |
Kruppel-like factor 2 |
3139 |
0.19 |
| chr6_72129092_72129273 | 0.13 |
ENSG00000207827 |
. |
15858 |
0.22 |
| chr13_31665227_31665378 | 0.13 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
47403 |
0.17 |
| chr10_121161636_121161837 | 0.13 |
GRK5 |
G protein-coupled receptor kinase 5 |
23787 |
0.17 |
| chr10_135334394_135334545 | 0.13 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
559 |
0.69 |
| chr16_30825894_30826089 | 0.13 |
ZNF629 |
zinc finger protein 629 |
27468 |
0.08 |
| chr11_122050907_122051108 | 0.13 |
ENSG00000207994 |
. |
27991 |
0.16 |
| chr22_24092778_24093143 | 0.13 |
ZNF70 |
zinc finger protein 70 |
319 |
0.77 |
| chr1_156116419_156116573 | 0.13 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
661 |
0.56 |
| chr16_2184293_2184536 | 0.13 |
ENSG00000265867 |
. |
1294 |
0.14 |
| chr10_29977190_29977386 | 0.13 |
ENSG00000222092 |
. |
23556 |
0.21 |
| chr10_79291588_79291953 | 0.13 |
ENSG00000199592 |
. |
55037 |
0.15 |
| chr22_23799075_23799226 | 0.13 |
ZDHHC8P1 |
zinc finger, DHHC-type containing 8 pseudogene 1 |
54806 |
0.11 |
| chr9_139221044_139221275 | 0.12 |
GPSM1 |
G-protein signaling modulator 1 |
773 |
0.48 |
| chrY_20310575_20310758 | 0.12 |
ENSG00000252166 |
. |
31932 |
0.23 |
| chr19_6067548_6067787 | 0.12 |
CTC-232P5.3 |
|
297 |
0.87 |
| chr11_131487115_131487266 | 0.12 |
NTM |
neurotrimin |
35285 |
0.18 |
| chr16_31117201_31117391 | 0.12 |
BCKDK |
branched chain ketoacid dehydrogenase kinase |
132 |
0.87 |
| chr10_126916601_126916752 | 0.12 |
CTBP2 |
C-terminal binding protein 2 |
67046 |
0.13 |
| chr2_97466776_97466927 | 0.12 |
ENSG00000264157 |
. |
2836 |
0.19 |
| chr1_23866282_23866470 | 0.12 |
E2F2 |
E2F transcription factor 2 |
8664 |
0.18 |
| chr12_110510869_110511513 | 0.12 |
C12orf76 |
chromosome 12 open reading frame 76 |
300 |
0.91 |
| chr6_151188569_151188720 | 0.12 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
951 |
0.67 |
| chr19_58350280_58350431 | 0.12 |
ZNF587B |
zinc finger protein 587B |
8674 |
0.1 |
| chr6_80175862_80176013 | 0.12 |
LCA5 |
Leber congenital amaurosis 5 |
71188 |
0.12 |
| chr3_156272520_156272859 | 0.12 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
55 |
0.98 |
| chr7_5517932_5518149 | 0.12 |
ENSG00000238394 |
. |
6986 |
0.13 |
| chr11_15601328_15601479 | 0.12 |
ENSG00000253072 |
. |
97964 |
0.09 |
| chrY_14774767_14775041 | 0.12 |
USP9Y |
ubiquitin specific peptidase 9, Y-linked |
38256 |
0.23 |
| chr10_89420631_89420782 | 0.12 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
2 |
0.96 |
| chr20_4707737_4707988 | 0.12 |
PRND |
prion protein 2 (dublet) |
5306 |
0.21 |
| chr2_242210722_242211682 | 0.12 |
HDLBP |
high density lipoprotein binding protein |
184 |
0.93 |
| chr5_32490035_32490410 | 0.12 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
41517 |
0.15 |
| chr17_42637074_42637426 | 0.12 |
FZD2 |
frizzled family receptor 2 |
2325 |
0.29 |
| chr4_17818884_17819094 | 0.12 |
NCAPG |
non-SMC condensin I complex, subunit G |
6333 |
0.21 |
| chr11_65194450_65194730 | 0.12 |
ENSG00000245532 |
. |
17339 |
0.11 |
| chr2_146281977_146282128 | 0.12 |
ENSG00000253036 |
. |
189414 |
0.03 |
| chr6_27463328_27463580 | 0.12 |
ZNF184 |
zinc finger protein 184 |
22557 |
0.21 |
| chr2_65215337_65216235 | 0.12 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
162 |
0.95 |
| chr1_53528292_53528443 | 0.12 |
PODN |
podocan |
482 |
0.76 |
| chr1_145576060_145576252 | 0.12 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
132 |
0.93 |
| chr4_80997222_80997373 | 0.12 |
ANTXR2 |
anthrax toxin receptor 2 |
2671 |
0.39 |
| chr7_91808687_91808971 | 0.12 |
LRRD1 |
leucine-rich repeats and death domain containing 1 |
16 |
0.97 |
| chr2_179142472_179142690 | 0.12 |
OSBPL6 |
oxysterol binding protein-like 6 |
7073 |
0.26 |
| chr10_92794688_92794839 | 0.11 |
ENSG00000201604 |
. |
112171 |
0.06 |
| chr2_87756158_87756657 | 0.11 |
RP11-1399P15.1 |
|
21146 |
0.27 |
| chr17_17380267_17380681 | 0.11 |
MED9 |
mediator complex subunit 9 |
166 |
0.94 |
| chr1_65400046_65400380 | 0.11 |
JAK1 |
Janus kinase 1 |
31974 |
0.2 |
| chr16_31084353_31084557 | 0.11 |
ZNF668 |
zinc finger protein 668 |
307 |
0.69 |
| chr14_103490453_103490773 | 0.11 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
33186 |
0.15 |
| chr14_23834178_23834353 | 0.11 |
EFS |
embryonal Fyn-associated substrate |
154 |
0.88 |
| chr9_95523893_95524044 | 0.11 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
3126 |
0.26 |
| chr13_36491458_36491923 | 0.11 |
DCLK1 |
doublecortin-like kinase 1 |
61911 |
0.15 |
| chr2_121650695_121650846 | 0.11 |
GLI2 |
GLI family zinc finger 2 |
95849 |
0.08 |
| chr2_105371997_105372148 | 0.11 |
ENSG00000207249 |
. |
22380 |
0.21 |
| chr12_68915355_68915540 | 0.11 |
RAP1B |
RAP1B, member of RAS oncogene family |
89172 |
0.08 |
| chr3_52566513_52566726 | 0.11 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
928 |
0.41 |
| chr17_66957491_66957642 | 0.11 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
6074 |
0.25 |
| chr2_97593731_97594362 | 0.11 |
ENSG00000252845 |
. |
28506 |
0.12 |
| chr11_68596728_68596879 | 0.11 |
CPT1A |
carnitine palmitoyltransferase 1A (liver) |
10349 |
0.2 |
| chr1_104112980_104113131 | 0.11 |
AMY2B |
amylase, alpha 2B (pancreatic) |
5402 |
0.19 |
| chr2_111230131_111230333 | 0.11 |
LIMS3 |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein |
161 |
0.49 |
| chr5_81065115_81065411 | 0.11 |
SSBP2 |
single-stranded DNA binding protein 2 |
18191 |
0.28 |
| chr13_99506197_99506352 | 0.11 |
DOCK9 |
dedicator of cytokinesis 9 |
1964 |
0.38 |
| chr3_40566463_40567204 | 0.11 |
ZNF621 |
zinc finger protein 621 |
74 |
0.97 |
| chr22_29427564_29428192 | 0.11 |
ZNRF3-AS1 |
ZNRF3 antisense RNA 1 |
414 |
0.8 |
| chrX_11162485_11162636 | 0.10 |
ENSG00000207151 |
. |
28247 |
0.2 |
| chr2_96915646_96915797 | 0.10 |
TMEM127 |
transmembrane protein 127 |
10612 |
0.14 |
| chr2_43144698_43145013 | 0.10 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
125123 |
0.05 |
| chr10_64893167_64893712 | 0.10 |
NRBF2 |
nuclear receptor binding factor 2 |
354 |
0.88 |
| chr10_30348565_30348787 | 0.10 |
KIAA1462 |
KIAA1462 |
223 |
0.96 |
| chr12_121570606_121571339 | 0.10 |
P2RX7 |
purinergic receptor P2X, ligand-gated ion channel, 7 |
198 |
0.94 |
| chr2_102803614_102803903 | 0.10 |
IL1RL2 |
interleukin 1 receptor-like 2 |
16 |
0.98 |
| chr3_105086242_105086708 | 0.10 |
ALCAM |
activated leukocyte cell adhesion molecule |
288 |
0.96 |
| chr15_58798000_58798151 | 0.10 |
RP11-355N15.1 |
|
7141 |
0.18 |
| chr8_82007271_82007422 | 0.10 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
16957 |
0.27 |
| chr3_75445432_75445834 | 0.10 |
ENSG00000266685 |
. |
27136 |
0.18 |
| chr4_173063385_173063536 | 0.10 |
GALNTL6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
89864 |
0.11 |
| chr1_7764600_7765026 | 0.10 |
CAMTA1 |
calmodulin binding transcription activator 1 |
31655 |
0.17 |
| chr20_3451275_3451544 | 0.10 |
ATRN |
attractin |
278 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.0 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.0 | GO:0071371 | cellular response to gonadotropin stimulus(GO:0071371) |
| 0.0 | 0.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.2 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |