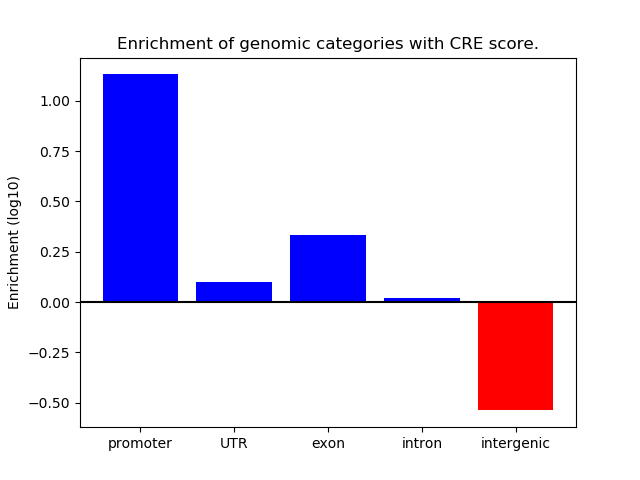
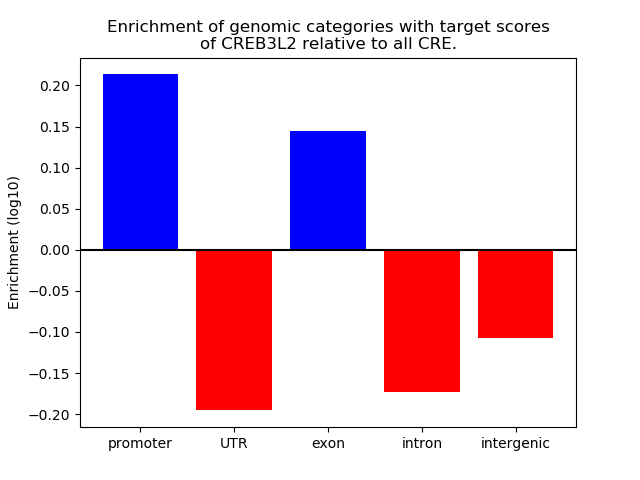
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
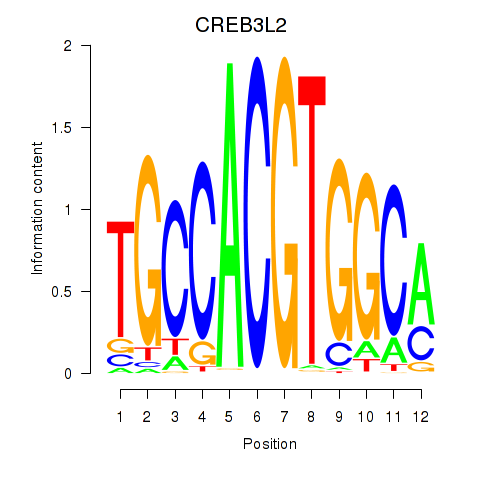
Results for CREB3L2
Z-value: 0.95
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.10 | cAMP responsive element binding protein 3 like 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_137671037_137671188 | CREB3L2 | 15681 | 0.188074 | 0.93 | 2.7e-04 | Click! |
| chr7_137683078_137683229 | CREB3L2 | 3640 | 0.246252 | 0.68 | 4.4e-02 | Click! |
| chr7_137665787_137665938 | CREB3L2 | 20931 | 0.177026 | 0.67 | 5.0e-02 | Click! |
| chr7_137671468_137671947 | CREB3L2 | 15086 | 0.189262 | 0.61 | 7.9e-02 | Click! |
| chr7_137685974_137686842 | CREB3L2 | 385 | 0.868788 | 0.59 | 9.6e-02 | Click! |
Activity of the CREB3L2 motif across conditions
Conditions sorted by the z-value of the CREB3L2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
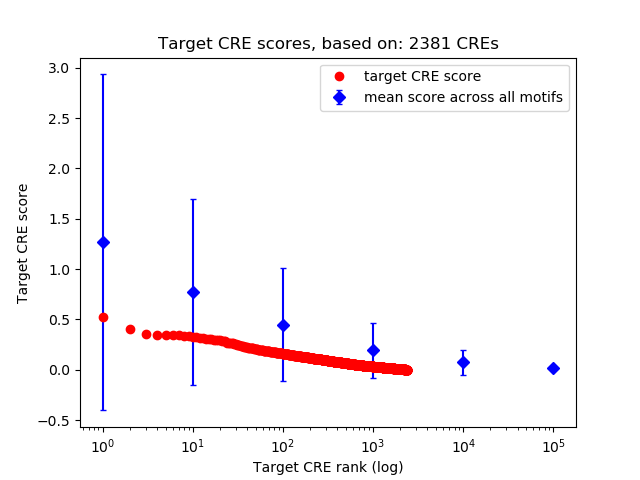
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_102106282_102106433 | 0.52 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
1339 |
0.56 |
| chr2_151337593_151337891 | 0.40 |
RND3 |
Rho family GTPase 3 |
4154 |
0.38 |
| chr2_234176635_234176786 | 0.35 |
ENSG00000252010 |
. |
7663 |
0.14 |
| chr2_27341761_27341954 | 0.34 |
CGREF1 |
cell growth regulator with EF-hand domain 1 |
41 |
0.94 |
| chr3_19988451_19988692 | 0.34 |
RAB5A |
RAB5A, member RAS oncogene family |
0 |
0.54 |
| chr1_118728319_118728470 | 0.34 |
SPAG17 |
sperm associated antigen 17 |
548 |
0.86 |
| chr13_24211833_24211984 | 0.34 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
58389 |
0.14 |
| chr3_29376770_29376984 | 0.33 |
ENSG00000216169 |
. |
34035 |
0.18 |
| chr15_96952055_96952206 | 0.33 |
AC087477.1 |
Uncharacterized protein |
47643 |
0.13 |
| chr9_95970167_95970323 | 0.33 |
WNK2 |
WNK lysine deficient protein kinase 2 |
23020 |
0.19 |
| chr5_178770702_178770853 | 0.32 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
1654 |
0.49 |
| chr22_44222171_44222754 | 0.32 |
RP3-388M5.9 |
|
14089 |
0.16 |
| chr3_123602332_123603013 | 0.31 |
MYLK |
myosin light chain kinase |
477 |
0.85 |
| chr17_47210248_47210565 | 0.31 |
B4GALNT2 |
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
77 |
0.97 |
| chr5_114795809_114796168 | 0.30 |
FEM1C |
fem-1 homolog c (C. elegans) |
84603 |
0.08 |
| chr4_120987081_120987232 | 0.30 |
RP11-679C8.2 |
|
957 |
0.45 |
| chr12_2185524_2185819 | 0.30 |
CACNA1C |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
22942 |
0.16 |
| chr12_105476938_105477089 | 0.30 |
ALDH1L2 |
aldehyde dehydrogenase 1 family, member L2 |
1328 |
0.39 |
| chr15_41575867_41576018 | 0.30 |
CHP1 |
calcineurin-like EF-hand protein 1 |
26834 |
0.13 |
| chr6_107816329_107816751 | 0.29 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
5378 |
0.23 |
| chr2_25934983_25935152 | 0.29 |
ENSG00000222070 |
. |
9994 |
0.19 |
| chr10_119303143_119303333 | 0.29 |
EMX2 |
empty spiracles homeobox 2 |
729 |
0.61 |
| chr5_154070973_154071336 | 0.28 |
ENSG00000221552 |
. |
5818 |
0.17 |
| chr2_241496900_241497119 | 0.27 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
370 |
0.78 |
| chrY_15864765_15864916 | 0.27 |
KALP |
Kallmann syndrome sequence pseudogene |
1304 |
0.55 |
| chrX_129245775_129245926 | 0.27 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
1159 |
0.52 |
| chr22_28181066_28181439 | 0.27 |
MN1 |
meningioma (disrupted in balanced translocation) 1 |
16234 |
0.25 |
| chr11_69460305_69460629 | 0.26 |
CCND1 |
cyclin D1 |
4493 |
0.24 |
| chr6_134211072_134211606 | 0.26 |
TCF21 |
transcription factor 21 |
866 |
0.5 |
| chr4_15481095_15481291 | 0.25 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
346 |
0.89 |
| chr6_147235316_147235787 | 0.25 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
1470 |
0.57 |
| chr8_17193981_17194132 | 0.25 |
MTMR7 |
myotubularin related protein 7 |
24874 |
0.21 |
| chr9_131006944_131007312 | 0.25 |
ENSG00000207581 |
. |
19 |
0.68 |
| chr10_5617288_5617517 | 0.24 |
CALML3-AS1 |
CALML3 antisense RNA 1 |
49193 |
0.09 |
| chr16_28213590_28213741 | 0.24 |
XPO6 |
exportin 6 |
9132 |
0.18 |
| chr4_157890038_157890662 | 0.23 |
PDGFC |
platelet derived growth factor C |
1705 |
0.41 |
| chr10_79261419_79261649 | 0.23 |
ENSG00000199592 |
. |
85273 |
0.09 |
| chr2_238316542_238316753 | 0.23 |
COL6A3 |
collagen, type VI, alpha 3 |
6144 |
0.22 |
| chr3_58037164_58037339 | 0.23 |
FLNB |
filamin B, beta |
26805 |
0.22 |
| chr21_46476184_46476390 | 0.22 |
AP001579.1 |
Uncharacterized protein |
16640 |
0.14 |
| chr20_19956104_19956326 | 0.22 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
41545 |
0.15 |
| chr12_77257769_77258091 | 0.22 |
RP11-461F16.3 |
|
14043 |
0.22 |
| chr3_30722457_30722608 | 0.22 |
RP11-1024P17.1 |
|
16621 |
0.27 |
| chr20_22559380_22559531 | 0.22 |
FOXA2 |
forkhead box A2 |
5646 |
0.34 |
| chr17_39472322_39472473 | 0.22 |
KRTAP17-1 |
keratin associated protein 17-1 |
450 |
0.64 |
| chr15_29396229_29396380 | 0.21 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
59777 |
0.14 |
| chr15_101629242_101629566 | 0.21 |
RP11-505E24.2 |
|
3133 |
0.3 |
| chr6_7910547_7910822 | 0.21 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
363 |
0.93 |
| chr9_126088906_126089175 | 0.21 |
CRB2 |
crumbs homolog 2 (Drosophila) |
29409 |
0.19 |
| chr20_50720613_50721041 | 0.21 |
ZFP64 |
ZFP64 zinc finger protein |
562 |
0.86 |
| chr10_128076215_128076453 | 0.21 |
ADAM12 |
ADAM metallopeptidase domain 12 |
690 |
0.79 |
| chr3_100708920_100709071 | 0.21 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
3302 |
0.32 |
| chr1_55792590_55792741 | 0.20 |
ENSG00000199831 |
. |
49860 |
0.14 |
| chr1_11791745_11791900 | 0.20 |
AGTRAP |
angiotensin II receptor-associated protein |
4319 |
0.14 |
| chr1_15653496_15653771 | 0.20 |
RP3-467K16.7 |
|
8327 |
0.16 |
| chr3_131221719_131221870 | 0.20 |
MRPL3 |
mitochondrial ribosomal protein L3 |
1 |
0.98 |
| chr13_52377985_52378296 | 0.20 |
DHRS12 |
dehydrogenase/reductase (SDR family) member 12 |
94 |
0.76 |
| chr5_159214342_159214493 | 0.19 |
ADRA1B |
adrenoceptor alpha 1B |
129373 |
0.05 |
| chr11_118691288_118691439 | 0.19 |
ENSG00000201535 |
. |
15356 |
0.12 |
| chr7_80928308_80928745 | 0.19 |
AC005008.2 |
Uncharacterized protein |
111420 |
0.08 |
| chr4_18023348_18023557 | 0.19 |
LCORL |
ligand dependent nuclear receptor corepressor-like |
31 |
0.99 |
| chr6_169475368_169475519 | 0.19 |
XXyac-YX65C7_A.2 |
|
137906 |
0.05 |
| chr3_183878422_183878793 | 0.19 |
DVL3 |
dishevelled segment polarity protein 3 |
4044 |
0.12 |
| chrX_139847263_139847516 | 0.19 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
19334 |
0.22 |
| chr1_65400046_65400380 | 0.19 |
JAK1 |
Janus kinase 1 |
31974 |
0.2 |
| chr7_100815791_100816003 | 0.19 |
NAT16 |
N-acetyltransferase 16 (GCN5-related, putative) |
6620 |
0.1 |
| chr1_247495331_247495594 | 0.19 |
ZNF496 |
zinc finger protein 496 |
417 |
0.84 |
| chr6_168558891_168559173 | 0.19 |
FRMD1 |
FERM domain containing 1 |
76795 |
0.11 |
| chr16_89368321_89368642 | 0.18 |
AC137932.5 |
|
4217 |
0.15 |
| chr3_169490145_169490517 | 0.18 |
MYNN |
myoneurin |
288 |
0.79 |
| chr19_35581704_35581880 | 0.18 |
AC020907.1 |
Uncharacterized protein |
15081 |
0.09 |
| chr7_116732017_116732168 | 0.18 |
ST7-AS2 |
ST7 antisense RNA 2 |
1258 |
0.49 |
| chr3_184078420_184078571 | 0.18 |
CLCN2 |
chloride channel, voltage-sensitive 2 |
895 |
0.31 |
| chr9_137437911_137438062 | 0.18 |
COL5A1 |
collagen, type V, alpha 1 |
95634 |
0.07 |
| chr17_33307453_33307681 | 0.18 |
LIG3 |
ligase III, DNA, ATP-dependent |
2 |
0.9 |
| chr4_48492552_48492703 | 0.18 |
ZAR1 |
zygote arrest 1 |
358 |
0.88 |
| chr14_73357781_73357932 | 0.18 |
DPF3 |
D4, zinc and double PHD fingers, family 3 |
2940 |
0.24 |
| chr10_100993963_100995023 | 0.18 |
HPSE2 |
heparanase 2 |
1066 |
0.66 |
| chr7_129194061_129194212 | 0.18 |
SMKR1 |
small lysine-rich protein 1 |
51503 |
0.12 |
| chr6_43970266_43970484 | 0.17 |
RP5-1120P11.1 |
|
1011 |
0.46 |
| chr18_52969288_52969439 | 0.17 |
TCF4 |
transcription factor 4 |
494 |
0.88 |
| chr15_34788505_34788693 | 0.17 |
ENSG00000221065 |
. |
31973 |
0.15 |
| chr17_38599985_38600404 | 0.17 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
481 |
0.74 |
| chr17_18061433_18061699 | 0.17 |
MYO15A |
myosin XVA |
56 |
0.96 |
| chr15_77713527_77713880 | 0.17 |
HMG20A |
high mobility group 20A |
350 |
0.83 |
| chr11_3077193_3077344 | 0.17 |
CARS |
cysteinyl-tRNA synthetase |
1364 |
0.33 |
| chr4_3306467_3306618 | 0.17 |
RGS12 |
regulator of G-protein signaling 12 |
4220 |
0.28 |
| chr6_18122528_18122892 | 0.17 |
NHLRC1 |
NHL repeat containing E3 ubiquitin protein ligase 1 |
141 |
0.96 |
| chr12_49318526_49318771 | 0.17 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
102 |
0.91 |
| chr9_19493377_19493724 | 0.17 |
RP11-363E7.4 |
|
40343 |
0.16 |
| chr12_120755596_120755815 | 0.17 |
ENSG00000252659 |
. |
4561 |
0.12 |
| chr2_198243514_198243752 | 0.17 |
SF3B1 |
splicing factor 3b, subunit 1, 155kDa |
20119 |
0.14 |
| chr7_100200956_100201118 | 0.17 |
PCOLCE-AS1 |
PCOLCE antisense RNA 1 |
379 |
0.65 |
| chr6_39760880_39761039 | 0.17 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
165 |
0.97 |
| chr17_66379730_66379881 | 0.16 |
ENSG00000207561 |
. |
40884 |
0.14 |
| chr2_217497534_217497685 | 0.16 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
58 |
0.97 |
| chrX_55187773_55187985 | 0.16 |
FAM104B |
family with sequence similarity 104, member B |
136 |
0.96 |
| chr7_23145115_23145340 | 0.16 |
KLHL7 |
kelch-like family member 7 |
126 |
0.97 |
| chr15_96877570_96877721 | 0.16 |
ENSG00000222651 |
. |
1155 |
0.34 |
| chr12_12447841_12447992 | 0.16 |
ENSG00000207010 |
. |
5653 |
0.19 |
| chr17_29887977_29888214 | 0.16 |
ENSG00000207614 |
. |
1080 |
0.35 |
| chr11_63774609_63774760 | 0.16 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
20370 |
0.1 |
| chr16_88450130_88450579 | 0.16 |
ZNF469 |
zinc finger protein 469 |
43525 |
0.15 |
| chr8_70668894_70669045 | 0.16 |
RP11-102F4.2 |
|
43344 |
0.14 |
| chr11_3043840_3044115 | 0.16 |
CARS-AS1 |
CARS antisense RNA 1 |
6647 |
0.13 |
| chr19_47447845_47447996 | 0.16 |
ENSG00000252071 |
. |
3626 |
0.22 |
| chr13_34361275_34361581 | 0.16 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
30758 |
0.21 |
| chr9_103494116_103494267 | 0.16 |
ENSG00000200966 |
. |
150530 |
0.04 |
| chr17_38603034_38603205 | 0.16 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
3406 |
0.18 |
| chrX_17394886_17395095 | 0.16 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
1447 |
0.46 |
| chr6_7672816_7672976 | 0.16 |
BMP6 |
bone morphogenetic protein 6 |
54134 |
0.14 |
| chr3_37493102_37493470 | 0.16 |
ITGA9 |
integrin, alpha 9 |
320 |
0.92 |
| chr5_179518071_179518222 | 0.16 |
RNF130 |
ring finger protein 130 |
19028 |
0.19 |
| chr19_51227387_51227538 | 0.15 |
CLEC11A |
C-type lectin domain family 11, member A |
682 |
0.53 |
| chr3_178789075_178789341 | 0.15 |
ZMAT3 |
zinc finger, matrin-type 3 |
146 |
0.96 |
| chr14_99848474_99848625 | 0.15 |
ENSG00000216173 |
. |
19921 |
0.19 |
| chr22_28174315_28174484 | 0.15 |
MN1 |
meningioma (disrupted in balanced translocation) 1 |
23087 |
0.23 |
| chr6_52422559_52422710 | 0.15 |
TRAM2 |
translocation associated membrane protein 2 |
19079 |
0.23 |
| chr17_1081882_1082496 | 0.15 |
ABR |
active BCR-related |
889 |
0.58 |
| chr11_94502592_94502995 | 0.15 |
AMOTL1 |
angiomotin like 1 |
1256 |
0.53 |
| chr1_2231502_2231862 | 0.15 |
RP4-713A8.1 |
|
26450 |
0.11 |
| chr6_137144069_137144462 | 0.15 |
PEX7 |
peroxisomal biogenesis factor 7 |
542 |
0.78 |
| chr9_126110255_126110406 | 0.15 |
CRB2 |
crumbs homolog 2 (Drosophila) |
8119 |
0.24 |
| chr7_101579659_101580126 | 0.15 |
CTB-181H17.1 |
|
23504 |
0.22 |
| chr1_28919066_28919537 | 0.15 |
RAB42 |
RAB42, member RAS oncogene family |
589 |
0.62 |
| chr19_41193747_41193898 | 0.15 |
NUMBL |
numb homolog (Drosophila)-like |
1675 |
0.25 |
| chr6_52437991_52438235 | 0.15 |
TRAM2 |
translocation associated membrane protein 2 |
3600 |
0.31 |
| chr22_36902639_36903091 | 0.14 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
204 |
0.92 |
| chr17_80008374_80008525 | 0.14 |
GPS1 |
G protein pathway suppressor 1 |
375 |
0.59 |
| chr21_36935983_36936169 | 0.14 |
ENSG00000211590 |
. |
156937 |
0.04 |
| chr1_244014741_244014953 | 0.14 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
466 |
0.87 |
| chr19_45826969_45827349 | 0.14 |
CKM |
creatine kinase, muscle |
924 |
0.44 |
| chr2_242626487_242626967 | 0.14 |
DTYMK |
deoxythymidylate kinase (thymidylate kinase) |
321 |
0.79 |
| chr12_110010896_110011058 | 0.14 |
MVK |
mevalonate kinase |
83 |
0.79 |
| chr10_74114795_74115085 | 0.14 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
33 |
0.98 |
| chr2_232328586_232328818 | 0.14 |
NCL |
nucleolin |
189 |
0.89 |
| chr12_109901272_109901510 | 0.14 |
KCTD10 |
potassium channel tetramerization domain containing 10 |
3191 |
0.21 |
| chr12_133613669_133614058 | 0.14 |
ZNF84 |
zinc finger protein 84 |
15 |
0.97 |
| chr15_90645383_90645632 | 0.14 |
IDH2 |
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
193 |
0.92 |
| chr6_110720825_110721164 | 0.14 |
DDO |
D-aspartate oxidase |
15771 |
0.18 |
| chr1_57890417_57890605 | 0.14 |
DAB1 |
Dab, reelin signal transducer, homolog 1 (Drosophila) |
782 |
0.79 |
| chr17_18537909_18538132 | 0.14 |
TBC1D28 |
TBC1 domain family, member 28 |
7717 |
0.13 |
| chr1_202896403_202896578 | 0.14 |
KLHL12 |
kelch-like family member 12 |
100 |
0.95 |
| chr3_39190203_39190389 | 0.14 |
CSRNP1 |
cysteine-serine-rich nuclear protein 1 |
4770 |
0.2 |
| chr19_41111211_41111409 | 0.14 |
ENSG00000266164 |
. |
1645 |
0.28 |
| chr16_70148022_70148173 | 0.14 |
PDPR |
pyruvate dehydrogenase phosphatase regulatory subunit |
178 |
0.94 |
| chr16_28985699_28985940 | 0.14 |
SPNS1 |
spinster homolog 1 (Drosophila) |
177 |
0.87 |
| chr6_136871727_136872192 | 0.13 |
MAP7 |
microtubule-associated protein 7 |
2 |
0.67 |
| chr3_49940383_49940534 | 0.13 |
MST1R |
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
584 |
0.48 |
| chrX_52964541_52964769 | 0.13 |
FAM156A |
family with sequence similarity 156, member A |
20874 |
0.17 |
| chr9_82278752_82279150 | 0.13 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
11443 |
0.33 |
| chr3_148709208_148709359 | 0.13 |
GYG1 |
glycogenin 1 |
16 |
0.98 |
| chr3_9958820_9959089 | 0.13 |
IL17RC |
interleukin 17 receptor C |
46 |
0.93 |
| chr12_65959878_65960118 | 0.13 |
MSRB3 |
methionine sulfoxide reductase B3 |
239343 |
0.02 |
| chr6_161664088_161664497 | 0.13 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
30759 |
0.25 |
| chr18_59537327_59537518 | 0.13 |
RNF152 |
ring finger protein 152 |
23570 |
0.27 |
| chrX_17395110_17395408 | 0.13 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
1716 |
0.4 |
| chr6_31802720_31802923 | 0.13 |
C6orf48 |
chromosome 6 open reading frame 48 |
112 |
0.61 |
| chr19_44007928_44008284 | 0.13 |
PHLDB3 |
pleckstrin homology-like domain, family B, member 3 |
460 |
0.71 |
| chr17_20812306_20812457 | 0.13 |
RP11-344E13.3 |
|
11977 |
0.13 |
| chr1_150254993_150255622 | 0.13 |
C1orf51 |
chromosome 1 open reading frame 51 |
78 |
0.94 |
| chr18_158238_158410 | 0.13 |
USP14 |
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
59 |
0.98 |
| chr18_53398919_53399070 | 0.13 |
TCF4 |
transcription factor 4 |
66976 |
0.14 |
| chr2_242823909_242824060 | 0.13 |
AC131097.3 |
|
115 |
0.94 |
| chr21_47038314_47038465 | 0.13 |
PCBP3 |
poly(rC) binding protein 3 |
25219 |
0.21 |
| chr13_97992448_97992599 | 0.13 |
MBNL2 |
muscleblind-like splicing regulator 2 |
16527 |
0.22 |
| chr3_192289269_192289452 | 0.13 |
ENSG00000238902 |
. |
37459 |
0.17 |
| chr1_68297532_68297957 | 0.13 |
GNG12-AS1 |
GNG12 antisense RNA 1 |
242 |
0.9 |
| chrX_292077_292313 | 0.13 |
LINC00685 |
long intergenic non-protein coding RNA 685 |
10470 |
0.23 |
| chr3_61668665_61668816 | 0.13 |
ENSG00000252420 |
. |
56163 |
0.17 |
| chr10_76994278_76994482 | 0.13 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
1303 |
0.47 |
| chr9_46168304_46168455 | 0.13 |
ENSG00000238413 |
. |
26704 |
0.23 |
| chr5_91833_92077 | 0.13 |
PLEKHG4B |
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
48418 |
0.13 |
| chr14_76446267_76446546 | 0.13 |
TGFB3 |
transforming growth factor, beta 3 |
930 |
0.62 |
| chr8_80681214_80681467 | 0.13 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
1242 |
0.56 |
| chr19_35939103_35939363 | 0.13 |
FFAR2 |
free fatty acid receptor 2 |
30 |
0.96 |
| chr16_839466_839674 | 0.13 |
CHTF18 |
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
257 |
0.75 |
| chr8_1771644_1772131 | 0.13 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
255 |
0.93 |
| chr14_26994857_26995008 | 0.13 |
RP11-483C6.1 |
|
68791 |
0.11 |
| chr3_72091935_72092086 | 0.12 |
ENSG00000239250 |
. |
215682 |
0.02 |
| chr1_214397865_214398571 | 0.12 |
SMYD2 |
SET and MYND domain containing 2 |
56358 |
0.16 |
| chr20_25208343_25208494 | 0.12 |
AL035252.1 |
HCG2018895; Uncharacterized protein |
1048 |
0.47 |
| chrX_129039705_129039949 | 0.12 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
270 |
0.92 |
| chr3_42697942_42698093 | 0.12 |
RP4-613B23.1 |
|
2137 |
0.17 |
| chrX_56790465_56790994 | 0.12 |
ENSG00000204272 |
. |
35037 |
0.23 |
| chr11_79718516_79718667 | 0.12 |
ENSG00000221551 |
. |
315168 |
0.01 |
| chr7_5637609_5637772 | 0.12 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
3373 |
0.21 |
| chr6_133562382_133562833 | 0.12 |
EYA4 |
eyes absent homolog 4 (Drosophila) |
51 |
0.99 |
| chr19_2783530_2783806 | 0.12 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
299 |
0.8 |
| chr7_150065813_150066064 | 0.12 |
REPIN1 |
replication initiator 1 |
22 |
0.52 |
| chr17_49230988_49231211 | 0.12 |
NME1 |
NME/NM23 nucleoside diphosphate kinase 1 |
73 |
0.37 |
| chr10_126359460_126359611 | 0.12 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
32659 |
0.18 |
| chr3_30691423_30691574 | 0.12 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
43405 |
0.18 |
| chr8_143782547_143782698 | 0.12 |
LY6K |
lymphocyte antigen 6 complex, locus K |
732 |
0.49 |
| chr22_30641304_30641455 | 0.12 |
RP1-102K2.8 |
|
605 |
0.52 |
| chr8_122652196_122652655 | 0.12 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
283 |
0.88 |
| chr8_102121369_102121647 | 0.12 |
ENSG00000202360 |
. |
28679 |
0.21 |
| chr5_141257083_141257234 | 0.12 |
PCDH1 |
protocadherin 1 |
796 |
0.64 |
| chr5_118733448_118733675 | 0.12 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
41831 |
0.13 |
| chr4_20256302_20256578 | 0.12 |
SLIT2 |
slit homolog 2 (Drosophila) |
103 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0090266 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0034227 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.2 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.1 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |