Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
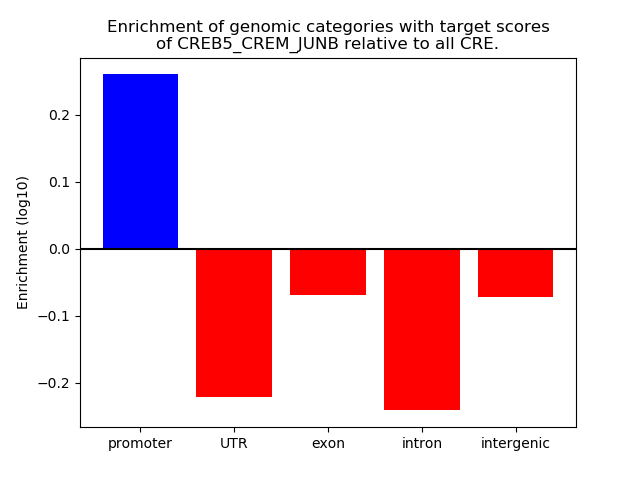
Results for CREB5_CREM_JUNB
Z-value: 0.60
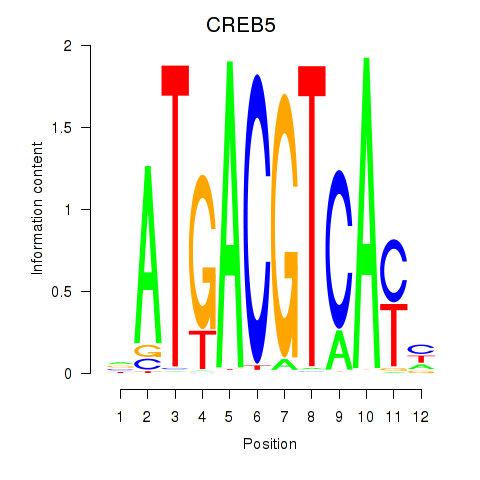
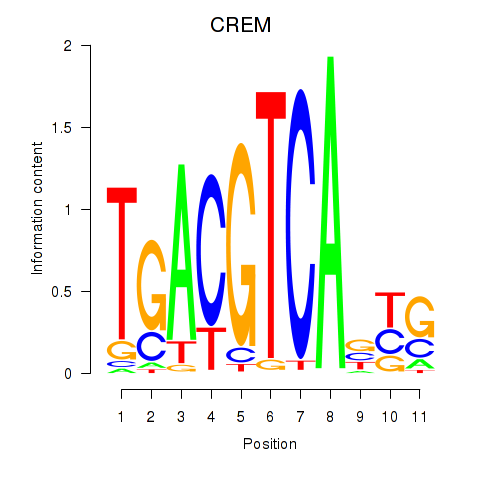
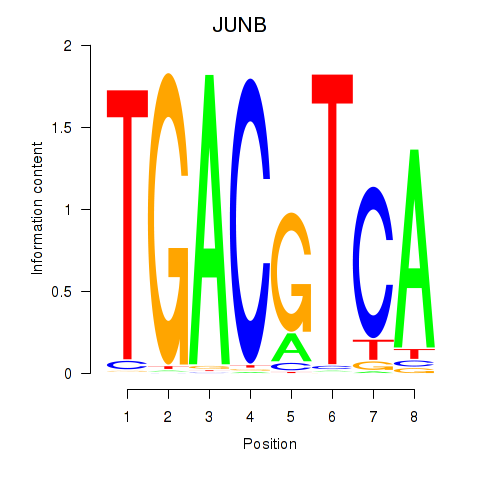
Transcription factors associated with CREB5_CREM_JUNB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB5
|
ENSG00000146592.12 | cAMP responsive element binding protein 5 |
|
CREM
|
ENSG00000095794.15 | cAMP responsive element modulator |
|
JUNB
|
ENSG00000171223.4 | JunB proto-oncogene, AP-1 transcription factor subunit |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_28831481_28831632 | CREB5 | 17470 | 0.283213 | 0.83 | 6.1e-03 | Click! |
| chr7_28527889_28528184 | CREB5 | 2712 | 0.426629 | 0.74 | 2.2e-02 | Click! |
| chr7_28520636_28520787 | CREB5 | 10037 | 0.317780 | -0.73 | 2.5e-02 | Click! |
| chr7_28893641_28893920 | CREB5 | 44754 | 0.182738 | 0.68 | 4.5e-02 | Click! |
| chr7_28705588_28705739 | CREB5 | 19935 | 0.287766 | 0.65 | 5.8e-02 | Click! |
| chr10_35473384_35473535 | CREM | 8828 | 0.152255 | -0.58 | 1.0e-01 | Click! |
| chr10_35415781_35416197 | CREM | 30 | 0.501216 | 0.56 | 1.2e-01 | Click! |
| chr10_35416363_35416802 | CREM | 177 | 0.833617 | 0.43 | 2.4e-01 | Click! |
| chr10_35484694_35485080 | CREM | 42 | 0.968921 | 0.40 | 2.8e-01 | Click! |
| chr10_35454329_35454480 | CREM | 2040 | 0.267658 | 0.35 | 3.5e-01 | Click! |
| chr19_12902481_12903520 | JUNB | 690 | 0.389467 | 0.84 | 5.0e-03 | Click! |
| chr19_12899780_12900482 | JUNB | 2179 | 0.106758 | 0.83 | 5.5e-03 | Click! |
| chr19_12903915_12904171 | JUNB | 1733 | 0.135908 | 0.55 | 1.2e-01 | Click! |
| chr19_12903527_12903836 | JUNB | 1371 | 0.175965 | 0.52 | 1.5e-01 | Click! |
| chr19_12901610_12901988 | JUNB | 511 | 0.517306 | 0.25 | 5.1e-01 | Click! |
Activity of the CREB5_CREM_JUNB motif across conditions
Conditions sorted by the z-value of the CREB5_CREM_JUNB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
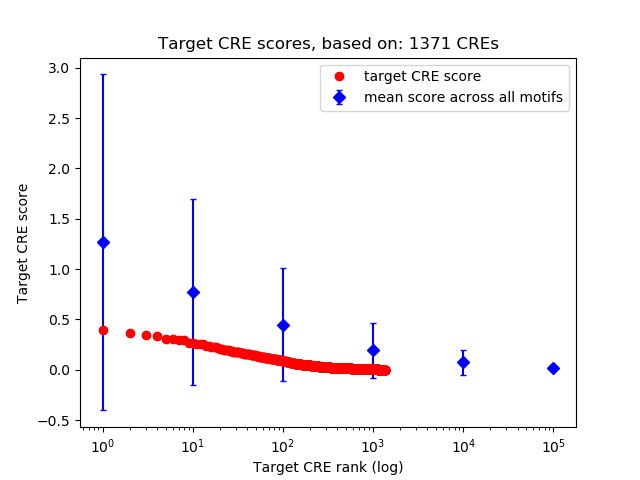
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_21967077_21967500 | 0.39 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
356 |
0.79 |
| chr20_16926810_16926961 | 0.37 |
ENSG00000212165 |
. |
36793 |
0.22 |
| chr5_176881613_176881764 | 0.35 |
PRR7 |
proline rich 7 (synaptic) |
6353 |
0.1 |
| chr5_170739718_170739894 | 0.34 |
TLX3 |
T-cell leukemia homeobox 3 |
3518 |
0.22 |
| chr4_15721295_15721446 | 0.31 |
BST1 |
bone marrow stromal cell antigen 1 |
4419 |
0.23 |
| chr12_56521312_56521715 | 0.30 |
ESYT1 |
extended synaptotagmin-like protein 1 |
327 |
0.64 |
| chr19_8454704_8454922 | 0.30 |
RAB11B |
RAB11B, member RAS oncogene family |
52 |
0.65 |
| chr17_4046800_4047053 | 0.30 |
CYB5D2 |
cytochrome b5 domain containing 2 |
70 |
0.9 |
| chr19_17666323_17666474 | 0.27 |
COLGALT1 |
collagen beta(1-O)galactosyltransferase 1 |
5 |
0.96 |
| chr6_91006489_91006667 | 0.26 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
49 |
0.98 |
| chr7_100808865_100809016 | 0.26 |
VGF |
VGF nerve growth factor inducible |
66 |
0.94 |
| chr15_59658629_59658780 | 0.26 |
FAM81A |
family with sequence similarity 81, member A |
6188 |
0.13 |
| chr16_86817661_86817812 | 0.26 |
FOXL1 |
forkhead box L1 |
205621 |
0.02 |
| chr2_8144573_8144775 | 0.24 |
ENSG00000221255 |
. |
427702 |
0.01 |
| chr2_28582064_28582304 | 0.24 |
FOSL2 |
FOS-like antigen 2 |
33485 |
0.14 |
| chr19_46009815_46010009 | 0.23 |
VASP |
vasodilator-stimulated phosphoprotein |
75 |
0.95 |
| chr12_71833497_71833776 | 0.22 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
86 |
0.97 |
| chr1_228356433_228356584 | 0.22 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
2992 |
0.14 |
| chr9_130860266_130860759 | 0.22 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
71 |
0.93 |
| chr12_54378782_54378933 | 0.21 |
HOXC10 |
homeobox C10 |
8 |
0.71 |
| chr17_2496882_2497033 | 0.21 |
PAFAH1B1 |
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
83 |
0.97 |
| chr9_137347969_137348661 | 0.20 |
RXRA |
retinoid X receptor, alpha |
49887 |
0.15 |
| chr11_18720267_18720541 | 0.20 |
TMEM86A |
transmembrane protein 86A |
66 |
0.96 |
| chr19_42900892_42901243 | 0.19 |
LIPE-AS1 |
LIPE antisense RNA 1 |
213 |
0.88 |
| chr3_170075366_170075517 | 0.19 |
SKIL |
SKI-like oncogene |
62 |
0.98 |
| chr1_228353838_228353989 | 0.19 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
397 |
0.6 |
| chr11_64044573_64044724 | 0.19 |
GPR137 |
G protein-coupled receptor 137 |
1488 |
0.15 |
| chr17_6926858_6927139 | 0.18 |
BCL6B |
B-cell CLL/lymphoma 6, member B |
202 |
0.83 |
| chr7_27232397_27232921 | 0.18 |
HOXA11-AS |
HOXA11 antisense RNA |
5221 |
0.07 |
| chr6_108486676_108487297 | 0.18 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
58 |
0.78 |
| chr19_55919228_55919426 | 0.18 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
182 |
0.86 |
| chr4_159131007_159131158 | 0.17 |
TMEM144 |
transmembrane protein 144 |
264 |
0.91 |
| chr1_155292308_155292690 | 0.17 |
RUSC1-AS1 |
RUSC1 antisense RNA 1 |
1164 |
0.22 |
| chr7_96634662_96635012 | 0.17 |
DLX6 |
distal-less homeobox 6 |
23 |
0.95 |
| chr17_7165860_7166195 | 0.17 |
CLDN7 |
claudin 7 |
230 |
0.78 |
| chr20_19915434_19915955 | 0.16 |
RIN2 |
Ras and Rab interactor 2 |
45484 |
0.16 |
| chr8_145555468_145555619 | 0.16 |
SCRT1 |
scratch family zinc finger 1 |
4400 |
0.08 |
| chr20_34203605_34204165 | 0.16 |
SPAG4 |
sperm associated antigen 4 |
71 |
0.95 |
| chr6_3157575_3157860 | 0.16 |
TUBB2A |
tubulin, beta 2A class IIa |
43 |
0.97 |
| chr5_58423468_58424016 | 0.16 |
RP11-266N13.2 |
|
88154 |
0.09 |
| chr7_38217813_38218059 | 0.16 |
STARD3NL |
STARD3 N-terminal like |
17 |
0.99 |
| chr19_55574631_55574782 | 0.16 |
RDH13 |
retinol dehydrogenase 13 (all-trans/9-cis) |
162 |
0.91 |
| chr20_15773685_15773836 | 0.15 |
MACROD2 |
MACRO domain containing 2 |
69690 |
0.14 |
| chr19_10974082_10974233 | 0.15 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
8032 |
0.12 |
| chr16_27378896_27379134 | 0.15 |
IL4R |
interleukin 4 receptor |
15019 |
0.19 |
| chr2_74781498_74781793 | 0.15 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
213 |
0.73 |
| chr16_12086577_12086728 | 0.15 |
RP11-166B2.7 |
|
14945 |
0.11 |
| chr19_51014778_51015051 | 0.15 |
JOSD2 |
Josephin domain containing 2 |
304 |
0.8 |
| chr17_18528902_18529084 | 0.14 |
CCDC144B |
coiled-coil domain containing 144B (pseudogene) |
72 |
0.96 |
| chr19_45349520_45349839 | 0.14 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
104 |
0.94 |
| chr16_31471330_31471687 | 0.14 |
ARMC5 |
armadillo repeat containing 5 |
226 |
0.67 |
| chr3_29228940_29229091 | 0.14 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
93458 |
0.09 |
| chr2_43822335_43823890 | 0.14 |
THADA |
thyroid adenoma associated |
1 |
0.98 |
| chr6_158508116_158508267 | 0.13 |
SYNJ2 |
synaptojanin 2 |
9479 |
0.2 |
| chr14_74352825_74353319 | 0.13 |
ZNF410 |
zinc finger protein 410 |
248 |
0.86 |
| chr9_137263893_137264044 | 0.13 |
ENSG00000263897 |
. |
7289 |
0.25 |
| chr16_85045097_85045598 | 0.13 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
206 |
0.95 |
| chr19_2956542_2956693 | 0.13 |
ZNF77 |
zinc finger protein 77 |
11686 |
0.12 |
| chr2_179278488_179279091 | 0.13 |
AC009948.5 |
|
39 |
0.97 |
| chr4_186316826_186317184 | 0.12 |
LRP2BP |
LRP2 binding protein |
48 |
0.69 |
| chr9_132250998_132251163 | 0.12 |
ENSG00000264298 |
. |
10245 |
0.21 |
| chr5_112849172_112849403 | 0.12 |
YTHDC2 |
YTH domain containing 2 |
93 |
0.97 |
| chr14_55738060_55738282 | 0.12 |
FBXO34 |
F-box protein 34 |
150 |
0.97 |
| chr11_14604750_14604901 | 0.12 |
PSMA1 |
proteasome (prosome, macropain) subunit, alpha type, 1 |
60338 |
0.12 |
| chr10_99078775_99079092 | 0.12 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
89 |
0.95 |
| chr5_107005011_107005570 | 0.12 |
EFNA5 |
ephrin-A5 |
1038 |
0.67 |
| chr17_4643181_4643351 | 0.12 |
CXCL16 |
chemokine (C-X-C motif) ligand 16 |
49 |
0.49 |
| chr11_108535331_108535645 | 0.12 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
264 |
0.95 |
| chr16_88859439_88859692 | 0.11 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
7946 |
0.09 |
| chr7_6388445_6388661 | 0.11 |
FAM220A |
family with sequence similarity 220, member A |
37 |
0.98 |
| chr8_135844464_135844657 | 0.11 |
ENSG00000199153 |
. |
27372 |
0.21 |
| chr3_8543267_8543418 | 0.11 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
1 |
0.52 |
| chr5_73730259_73730491 | 0.11 |
ENSG00000244326 |
. |
116564 |
0.06 |
| chr3_122513577_122513821 | 0.11 |
DIRC2 |
disrupted in renal carcinoma 2 |
57 |
0.95 |
| chr9_35617185_35617463 | 0.11 |
CD72 |
CD72 molecule |
1043 |
0.3 |
| chr12_57855074_57855466 | 0.11 |
GLI1 |
GLI family zinc finger 1 |
996 |
0.31 |
| chr16_31142045_31142196 | 0.11 |
RP11-388M20.2 |
|
634 |
0.42 |
| chr14_103988458_103988873 | 0.11 |
CKB |
creatine kinase, brain |
368 |
0.73 |
| chr22_36519910_36520146 | 0.11 |
APOL3 |
apolipoprotein L, 3 |
36803 |
0.17 |
| chr10_105728102_105728505 | 0.10 |
SLK |
STE20-like kinase |
1344 |
0.4 |
| chr1_63787954_63788471 | 0.10 |
RP4-792G4.2 |
|
83 |
0.88 |
| chr18_74205501_74206173 | 0.10 |
ZNF516 |
zinc finger protein 516 |
1309 |
0.32 |
| chr22_51059513_51059819 | 0.10 |
ARSA |
arylsulfatase A |
6917 |
0.1 |
| chr18_2654684_2655529 | 0.10 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
631 |
0.67 |
| chr17_79093601_79093928 | 0.10 |
ENSG00000207736 |
. |
5409 |
0.1 |
| chr21_44597084_44597458 | 0.10 |
CRYAA |
crystallin, alpha A |
7005 |
0.17 |
| chr8_33422545_33422696 | 0.10 |
RNF122 |
ring finger protein 122 |
2023 |
0.25 |
| chr8_13129460_13129611 | 0.10 |
DLC1 |
deleted in liver cancer 1 |
4520 |
0.3 |
| chr2_78182087_78182238 | 0.10 |
LRRTM4 |
leucine rich repeat transmembrane neuronal 4 |
361840 |
0.01 |
| chr7_993167_993948 | 0.10 |
ADAP1 |
ArfGAP with dual PH domains 1 |
776 |
0.55 |
| chr12_7282858_7283009 | 0.10 |
RP11-273B20.1 |
|
85 |
0.56 |
| chr3_33759550_33759763 | 0.10 |
CLASP2 |
cytoplasmic linker associated protein 2 |
115 |
0.98 |
| chr14_69260124_69260503 | 0.10 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
356 |
0.9 |
| chr17_55333458_55333748 | 0.10 |
MSI2 |
musashi RNA-binding protein 2 |
292 |
0.94 |
| chr10_33798417_33798757 | 0.09 |
NRP1 |
neuropilin 1 |
173397 |
0.03 |
| chr17_41281855_41282006 | 0.09 |
NBR2 |
neighbor of BRCA1 gene 2 (non-protein coding) |
1295 |
0.35 |
| chr2_64957643_64958616 | 0.09 |
ENSG00000253082 |
. |
45109 |
0.14 |
| chr12_7068434_7068709 | 0.09 |
ENSG00000207713 |
. |
4291 |
0.07 |
| chr17_19367994_19368145 | 0.09 |
ENSG00000265335 |
. |
44477 |
0.09 |
| chr9_133788553_133789184 | 0.09 |
FIBCD1 |
fibrinogen C domain containing 1 |
16498 |
0.16 |
| chr14_71353925_71354621 | 0.09 |
PCNX |
pecanex homolog (Drosophila) |
19849 |
0.26 |
| chr21_45209367_45209684 | 0.08 |
RRP1 |
ribosomal RNA processing 1 |
131 |
0.95 |
| chr20_56284812_56285148 | 0.08 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
22 |
0.99 |
| chr11_1404301_1404651 | 0.08 |
BRSK2 |
BR serine/threonine kinase 2 |
6653 |
0.2 |
| chr17_1953560_1953783 | 0.08 |
ENSG00000267195 |
. |
3 |
0.83 |
| chr22_42228451_42228869 | 0.08 |
SREBF2 |
sterol regulatory element binding transcription factor 2 |
449 |
0.71 |
| chr4_37686725_37687086 | 0.08 |
RELL1 |
RELT-like 1 |
1093 |
0.57 |
| chr1_155164087_155164666 | 0.08 |
ENSG00000271748 |
. |
592 |
0.39 |
| chr19_54960345_54960655 | 0.08 |
LENG8 |
leukocyte receptor cluster (LRC) member 8 |
94 |
0.72 |
| chr1_17630726_17630943 | 0.08 |
PADI4 |
peptidyl arginine deiminase, type IV |
3856 |
0.2 |
| chr3_52321810_52322234 | 0.08 |
WDR82 |
WD repeat domain 82 |
14 |
0.64 |
| chr10_16870022_16870173 | 0.08 |
RSU1 |
Ras suppressor protein 1 |
10570 |
0.28 |
| chr10_35484694_35485080 | 0.08 |
CREM |
cAMP responsive element modulator |
42 |
0.97 |
| chr1_29213318_29213476 | 0.08 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
206 |
0.93 |
| chr15_89905376_89905703 | 0.08 |
ENSG00000207819 |
. |
5709 |
0.19 |
| chr6_159519573_159519904 | 0.08 |
TAGAP |
T-cell activation RhoGTPase activating protein |
53554 |
0.11 |
| chr2_138880085_138880236 | 0.08 |
HNMT |
histamine N-methyltransferase |
158117 |
0.04 |
| chr20_56293559_56293931 | 0.07 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
7204 |
0.28 |
| chr7_4764745_4765032 | 0.07 |
FOXK1 |
forkhead box K1 |
15602 |
0.18 |
| chr16_67564286_67564635 | 0.07 |
FAM65A |
family with sequence similarity 65, member A |
766 |
0.44 |
| chr11_118122819_118123419 | 0.07 |
MPZL3 |
myelin protein zero-like 3 |
54 |
0.96 |
| chr20_12990547_12990774 | 0.07 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
663 |
0.83 |
| chr19_39175153_39175304 | 0.07 |
CTB-186G2.4 |
|
9018 |
0.12 |
| chr11_12989815_12989966 | 0.07 |
ENSG00000266625 |
. |
4703 |
0.25 |
| chr15_70250319_70250470 | 0.07 |
ENSG00000207965 |
. |
121413 |
0.06 |
| chr22_39745318_39745552 | 0.07 |
SYNGR1 |
synaptogyrin 1 |
495 |
0.71 |
| chr19_46931946_46932393 | 0.07 |
CCDC8 |
coiled-coil domain containing 8 |
15328 |
0.13 |
| chr11_33183160_33183580 | 0.07 |
CSTF3-AS1 |
CSTF3 antisense RNA 1 (head to head) |
136 |
0.7 |
| chr1_91183266_91183570 | 0.07 |
BARHL2 |
BarH-like homeobox 2 |
624 |
0.83 |
| chr19_1855668_1855910 | 0.06 |
CTB-31O20.6 |
|
3391 |
0.1 |
| chr16_2563812_2564354 | 0.06 |
ATP6V0C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
106 |
0.49 |
| chr12_66347998_66348149 | 0.06 |
RP11-366L20.3 |
|
5814 |
0.22 |
| chr6_157007333_157008165 | 0.06 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
91314 |
0.08 |
| chr10_71991819_71992862 | 0.06 |
PPA1 |
pyrophosphatase (inorganic) 1 |
820 |
0.66 |
| chr18_20840698_20840849 | 0.06 |
RP11-17J14.2 |
|
455 |
0.88 |
| chr19_36036432_36037158 | 0.06 |
AD000090.2 |
|
136 |
0.59 |
| chr11_82782830_82782981 | 0.06 |
RAB30 |
RAB30, member RAS oncogene family |
3 |
0.98 |
| chr17_17586273_17586527 | 0.06 |
RAI1 |
retinoic acid induced 1 |
591 |
0.75 |
| chr7_140396423_140396944 | 0.06 |
NDUFB2 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
105 |
0.63 |
| chr8_142275255_142276084 | 0.06 |
RP11-10J21.3 |
Uncharacterized protein |
11005 |
0.14 |
| chr10_14880634_14881306 | 0.06 |
CDNF |
cerebral dopamine neurotrophic factor |
396 |
0.57 |
| chrX_39759626_39759841 | 0.06 |
ENSG00000263972 |
. |
62918 |
0.14 |
| chr8_41605528_41605679 | 0.06 |
ANK1 |
ankyrin 1, erythrocytic |
41906 |
0.13 |
| chr3_51720796_51721049 | 0.06 |
ENSG00000201595 |
. |
7676 |
0.15 |
| chr1_28655191_28655470 | 0.06 |
MED18 |
mediator complex subunit 18 |
219 |
0.92 |
| chr17_61851142_61851414 | 0.06 |
DDX42 |
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
34 |
0.63 |
| chr2_190140835_190141232 | 0.06 |
ENSG00000266817 |
. |
35454 |
0.2 |
| chr8_39043803_39043954 | 0.06 |
ADAM32 |
ADAM metallopeptidase domain 32 |
78403 |
0.1 |
| chr8_145561219_145561461 | 0.06 |
SCRT1 |
scratch family zinc finger 1 |
1397 |
0.19 |
| chr9_100565675_100565842 | 0.06 |
FOXE1 |
forkhead box E1 (thyroid transcription factor 2) |
49778 |
0.12 |
| chr19_9546259_9546565 | 0.06 |
ZNF266 |
zinc finger protein 266 |
178 |
0.95 |
| chr17_46684210_46684361 | 0.06 |
HOXB6 |
homeobox B6 |
1931 |
0.13 |
| chr1_244013782_244014025 | 0.05 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
473 |
0.87 |
| chr4_5758904_5759055 | 0.05 |
EVC |
Ellis van Creveld syndrome |
46037 |
0.16 |
| chr2_238609518_238609709 | 0.05 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
8631 |
0.24 |
| chr16_81514485_81514779 | 0.05 |
CMIP |
c-Maf inducing protein |
14322 |
0.26 |
| chr9_19472304_19472455 | 0.05 |
RP11-363E7.4 |
|
19172 |
0.2 |
| chr20_17876219_17876496 | 0.05 |
ENSG00000212555 |
. |
8049 |
0.2 |
| chr13_24742029_24742180 | 0.05 |
ENSG00000252695 |
. |
5549 |
0.22 |
| chr8_142180071_142180222 | 0.05 |
DENND3 |
DENN/MADD domain containing 3 |
8074 |
0.2 |
| chr3_185080934_185081796 | 0.05 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
457 |
0.87 |
| chr6_47445012_47445717 | 0.05 |
CD2AP |
CD2-associated protein |
161 |
0.96 |
| chr1_226189668_226189925 | 0.05 |
SDE2 |
SDE2 telomere maintenance homolog (S. pombe) |
2764 |
0.23 |
| chr9_99851520_99851671 | 0.05 |
CTSV |
cathepsin V |
49670 |
0.15 |
| chr15_48858385_48858541 | 0.05 |
FBN1 |
fibrillin 1 |
79455 |
0.1 |
| chr16_87469465_87469755 | 0.05 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
2870 |
0.22 |
| chr7_106064692_106064843 | 0.05 |
CTB-111H14.1 |
|
80518 |
0.1 |
| chr16_68056878_68057148 | 0.05 |
DUS2 |
dihydrouridine synthase 2 |
143 |
0.85 |
| chr8_38243294_38243670 | 0.05 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
243 |
0.88 |
| chr16_55401499_55401650 | 0.05 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
22038 |
0.18 |
| chr4_174443030_174443487 | 0.05 |
HAND2-AS1 |
HAND2 antisense RNA 1 (head to head) |
5163 |
0.23 |
| chr3_50297522_50298009 | 0.05 |
SEMA3B-AS1 |
SEMA3B antisense RNA 1 (head to head) |
7038 |
0.09 |
| chr6_27862660_27863352 | 0.05 |
HIST1H2BO |
histone cluster 1, H2bo |
1803 |
0.13 |
| chr10_30744625_30744776 | 0.05 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
16949 |
0.22 |
| chr1_1490221_1490372 | 0.05 |
TMEM240 |
transmembrane protein 240 |
14463 |
0.09 |
| chr19_38908689_38908861 | 0.05 |
RASGRP4 |
RAS guanyl releasing protein 4 |
8027 |
0.09 |
| chr6_126660953_126661204 | 0.05 |
CENPW |
centromere protein W |
242 |
0.96 |
| chr7_1979405_1979706 | 0.05 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
630 |
0.8 |
| chr12_54411200_54411679 | 0.05 |
HOXC4 |
homeobox C4 |
724 |
0.35 |
| chr1_32205139_32205290 | 0.05 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
5119 |
0.16 |
| chr19_6393244_6393475 | 0.05 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
112 |
0.91 |
| chr16_70456934_70457085 | 0.05 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
15982 |
0.11 |
| chr9_111773050_111773591 | 0.05 |
CTNNAL1 |
catenin (cadherin-associated protein), alpha-like 1 |
2459 |
0.23 |
| chr2_160471179_160471674 | 0.05 |
AC009506.1 |
|
381 |
0.66 |
| chr2_108994212_108994393 | 0.05 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
166 |
0.96 |
| chrX_98854100_98854251 | 0.05 |
ENSG00000252430 |
. |
213901 |
0.03 |
| chr22_22020368_22020729 | 0.05 |
PPIL2 |
peptidylprolyl isomerase (cyclophilin)-like 2 |
190 |
0.88 |
| chr5_54070623_54070885 | 0.05 |
ENSG00000221073 |
. |
78022 |
0.1 |
| chr1_22379888_22380679 | 0.05 |
CDC42 |
cell division cycle 42 |
493 |
0.71 |
| chr19_5339568_5339818 | 0.05 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
1121 |
0.59 |
| chr19_54475084_54475235 | 0.04 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
8865 |
0.1 |
| chr11_61584512_61584663 | 0.04 |
FADS1 |
fatty acid desaturase 1 |
47 |
0.89 |
| chr11_119599926_119600166 | 0.04 |
CTD-2523D13.2 |
|
247 |
0.49 |
| chr17_41737027_41737178 | 0.04 |
MEOX1 |
mesenchyme homeobox 1 |
1829 |
0.33 |
| chr2_203735613_203735883 | 0.04 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
21 |
0.98 |
| chr11_64546452_64546665 | 0.04 |
SF1 |
splicing factor 1 |
300 |
0.83 |
| chr3_71478030_71478538 | 0.04 |
ENSG00000221264 |
. |
112956 |
0.06 |
| chr17_47841796_47842381 | 0.04 |
FAM117A |
family with sequence similarity 117, member A |
559 |
0.72 |
| chr8_146012278_146012963 | 0.04 |
ZNF34 |
zinc finger protein 34 |
69 |
0.94 |
| chr12_18876500_18876651 | 0.04 |
PLCZ1 |
phospholipase C, zeta 1 |
4028 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |