Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
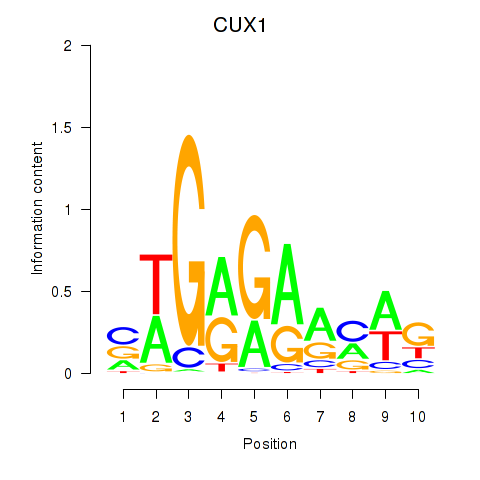
Results for CUX1
Z-value: 1.42
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.5 | cut like homeobox 1 |
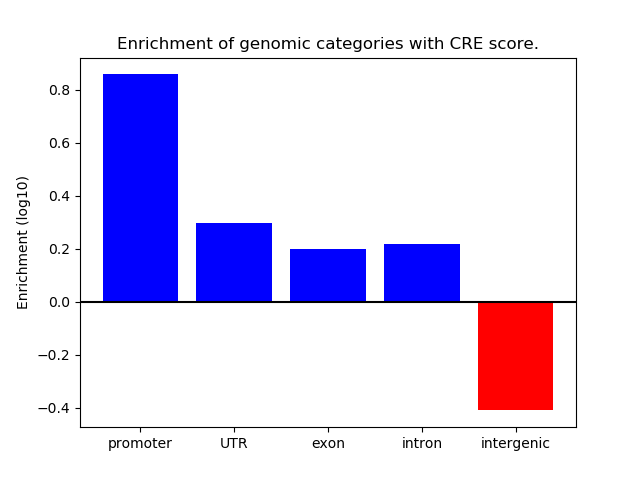
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_101460259_101460698 | CUX1 | 404 | 0.873916 | -0.77 | 1.5e-02 | Click! |
| chr7_101459490_101459645 | CUX1 | 266 | 0.931579 | -0.76 | 1.7e-02 | Click! |
| chr7_101459208_101459407 | CUX1 | 6 | 0.982319 | -0.75 | 2.0e-02 | Click! |
| chr7_101397584_101398512 | CUX1 | 60911 | 0.121573 | -0.61 | 8.3e-02 | Click! |
| chr7_101457714_101458422 | CUX1 | 891 | 0.644913 | -0.58 | 9.8e-02 | Click! |
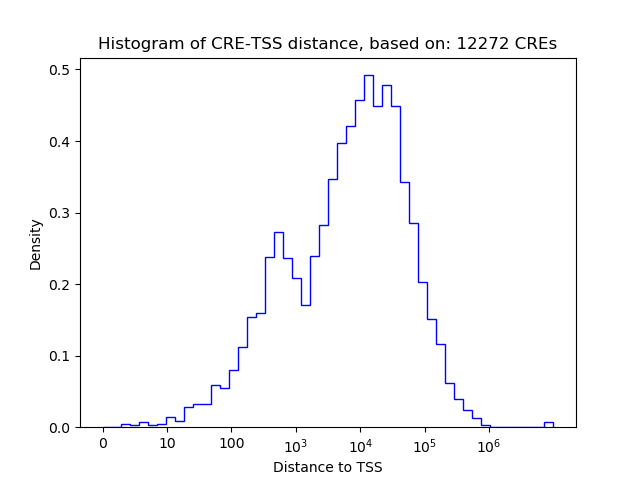
Activity of the CUX1 motif across conditions
Conditions sorted by the z-value of the CUX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
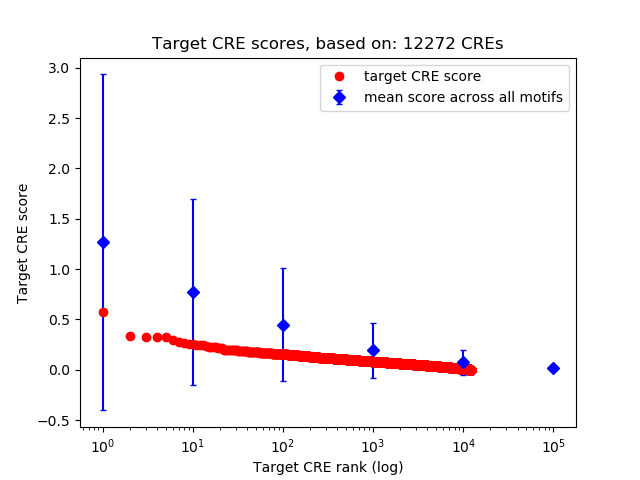
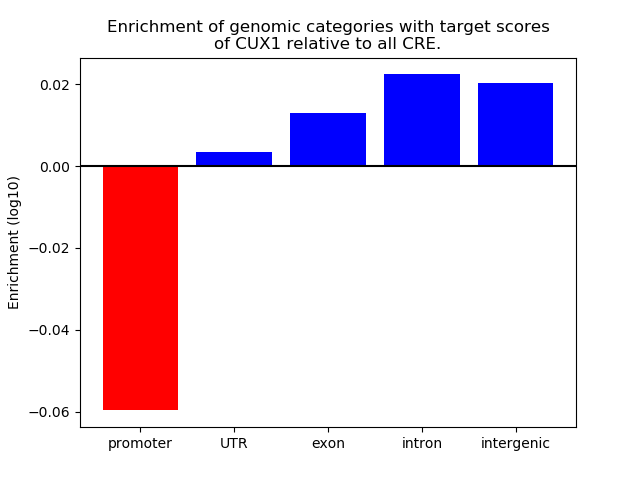
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_69674453_69675874 | 0.58 |
DLG3 |
discs, large homolog 3 (Drosophila) |
217 |
0.75 |
| chrX_39964334_39965624 | 0.34 |
BCOR |
BCL6 corepressor |
8323 |
0.32 |
| chr1_161038546_161039545 | 0.33 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr9_93577957_93578337 | 0.32 |
SYK |
spleen tyrosine kinase |
11623 |
0.32 |
| chr13_113597266_113598290 | 0.32 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
24979 |
0.15 |
| chr22_29601932_29602692 | 0.30 |
EMID1 |
EMI domain containing 1 |
400 |
0.8 |
| chr7_67384045_67384316 | 0.27 |
ENSG00000265600 |
. |
562513 |
0.0 |
| chr8_68304896_68305241 | 0.27 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
49156 |
0.18 |
| chr5_1110808_1112337 | 0.26 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
578 |
0.75 |
| chr1_245951382_245951568 | 0.26 |
RP11-522M21.3 |
|
111695 |
0.07 |
| chr16_28506747_28507012 | 0.25 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
24 |
0.94 |
| chr9_92275535_92275686 | 0.25 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
55657 |
0.16 |
| chr6_170604225_170605407 | 0.24 |
FAM120B |
family with sequence similarity 120B |
407 |
0.83 |
| chr10_101088892_101089952 | 0.24 |
CNNM1 |
cyclin M1 |
266 |
0.95 |
| chr1_22345507_22345745 | 0.23 |
ENSG00000201273 |
. |
8148 |
0.1 |
| chr11_14665333_14666638 | 0.23 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
608 |
0.56 |
| chr2_238607001_238607152 | 0.23 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
6094 |
0.25 |
| chr6_54711000_54712080 | 0.22 |
FAM83B |
family with sequence similarity 83, member B |
29 |
0.98 |
| chr20_52492319_52492470 | 0.22 |
AC005220.3 |
|
64305 |
0.13 |
| chr13_33589965_33590391 | 0.22 |
KL |
klotho |
29 |
0.98 |
| chr3_150887706_150887956 | 0.21 |
ENSG00000199994 |
. |
18055 |
0.16 |
| chr17_57408824_57410302 | 0.20 |
YPEL2 |
yippee-like 2 (Drosophila) |
500 |
0.78 |
| chr17_21279729_21280949 | 0.20 |
KCNJ12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
830 |
0.69 |
| chr13_113237033_113237219 | 0.20 |
TUBGCP3 |
tubulin, gamma complex associated protein 3 |
5313 |
0.28 |
| chr16_89766857_89768141 | 0.20 |
SPATA2L |
spermatogenesis associated 2-like |
549 |
0.59 |
| chr21_47344013_47344164 | 0.20 |
PRED62 |
Uncharacterized protein |
8389 |
0.2 |
| chr20_39122695_39122846 | 0.20 |
ENSG00000252434 |
. |
77255 |
0.12 |
| chr1_192130669_192130880 | 0.19 |
RGS18 |
regulator of G-protein signaling 18 |
3187 |
0.4 |
| chr12_14731074_14731371 | 0.19 |
PLBD1 |
phospholipase B domain containing 1 |
9939 |
0.15 |
| chr2_16615403_16615554 | 0.19 |
AC104623.2 |
|
88966 |
0.1 |
| chr2_103042024_103042175 | 0.19 |
IL18RAP |
interleukin 18 receptor accessory protein |
5953 |
0.15 |
| chr22_31613545_31613991 | 0.19 |
ENSG00000199695 |
. |
5071 |
0.12 |
| chr6_20404021_20405019 | 0.19 |
E2F3 |
E2F transcription factor 3 |
486 |
0.77 |
| chr10_106085822_106086336 | 0.19 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
2603 |
0.2 |
| chr20_3293982_3294133 | 0.19 |
C20orf194 |
chromosome 20 open reading frame 194 |
27551 |
0.12 |
| chr4_38602953_38603104 | 0.19 |
RP11-617D20.1 |
|
23168 |
0.2 |
| chr12_12735789_12735940 | 0.18 |
DUSP16 |
dual specificity phosphatase 16 |
20547 |
0.19 |
| chrX_152599728_152600567 | 0.18 |
ZNF275 |
zinc finger protein 275 |
534 |
0.76 |
| chr15_94841194_94841722 | 0.18 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
28 |
0.99 |
| chr2_74691535_74692741 | 0.18 |
MOGS |
mannosyl-oligosaccharide glucosidase |
351 |
0.67 |
| chr22_30577376_30577527 | 0.18 |
RP3-438O4.4 |
|
25647 |
0.13 |
| chr9_21964953_21966022 | 0.18 |
C9orf53 |
chromosome 9 open reading frame 53 |
1650 |
0.31 |
| chr22_41843038_41844299 | 0.18 |
TOB2 |
transducer of ERBB2, 2 |
641 |
0.64 |
| chr4_97275136_97275287 | 0.18 |
ENSG00000201640 |
. |
5022 |
0.36 |
| chr22_17577263_17577414 | 0.18 |
IL17RA |
interleukin 17 receptor A |
11489 |
0.15 |
| chr16_127905_128996 | 0.18 |
MPG |
N-methylpurine-DNA glycosylase |
194 |
0.87 |
| chr3_112686974_112687125 | 0.18 |
CD200R1 |
CD200 receptor 1 |
6710 |
0.17 |
| chr17_55889172_55889323 | 0.18 |
ENSG00000222976 |
. |
22437 |
0.16 |
| chr8_97352110_97352386 | 0.18 |
ENSG00000199732 |
. |
30698 |
0.17 |
| chr1_108492584_108492735 | 0.17 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
14406 |
0.21 |
| chrX_67913478_67914353 | 0.17 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
422 |
0.91 |
| chr1_204962096_204962247 | 0.17 |
NFASC |
neurofascin |
4141 |
0.24 |
| chr7_64461546_64461954 | 0.17 |
ERV3-1 |
endogenous retrovirus group 3, member 1 |
5281 |
0.17 |
| chr17_72740646_72740797 | 0.17 |
ENSG00000264624 |
. |
4031 |
0.11 |
| chr19_54714075_54714317 | 0.17 |
RPS9 |
ribosomal protein S9 |
9168 |
0.07 |
| chr12_107153685_107153836 | 0.17 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
14613 |
0.17 |
| chr1_181068882_181069172 | 0.17 |
IER5 |
immediate early response 5 |
11389 |
0.23 |
| chr13_27335081_27335232 | 0.17 |
GPR12 |
G protein-coupled receptor 12 |
234 |
0.96 |
| chr2_149894818_149895436 | 0.17 |
LYPD6B |
LY6/PLAUR domain containing 6B |
105 |
0.98 |
| chr3_50395480_50396917 | 0.17 |
TMEM115 |
transmembrane protein 115 |
843 |
0.33 |
| chr19_13906290_13907522 | 0.17 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
632 |
0.39 |
| chr17_41790734_41791669 | 0.17 |
ENSG00000215964 |
. |
34367 |
0.11 |
| chr3_71829345_71829567 | 0.17 |
PROK2 |
prokineticin 2 |
4756 |
0.22 |
| chr19_32836514_32837661 | 0.17 |
ZNF507 |
zinc finger protein 507 |
462 |
0.81 |
| chr3_46504181_46504332 | 0.17 |
LTF |
lactotransferrin |
905 |
0.54 |
| chr2_9142725_9143476 | 0.16 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
694 |
0.76 |
| chr16_72910024_72910217 | 0.16 |
ENSG00000251868 |
. |
54229 |
0.12 |
| chr2_63276750_63277690 | 0.16 |
OTX1 |
orthodenticle homeobox 1 |
28 |
0.97 |
| chr12_42862786_42862937 | 0.16 |
RP11-328C8.4 |
|
9693 |
0.15 |
| chr6_42504782_42504933 | 0.16 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
26943 |
0.17 |
| chr22_45604259_45604410 | 0.16 |
KIAA0930 |
KIAA0930 |
2604 |
0.24 |
| chr7_105898290_105898441 | 0.16 |
NAMPT |
nicotinamide phosphoribosyltransferase |
27002 |
0.18 |
| chr11_65627927_65628967 | 0.16 |
MUS81 |
MUS81 structure-specific endonuclease subunit |
89 |
0.82 |
| chr6_159476816_159477190 | 0.16 |
TAGAP |
T-cell activation RhoGTPase activating protein |
10819 |
0.2 |
| chr9_94184266_94184417 | 0.16 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
1803 |
0.49 |
| chr5_180631418_180632621 | 0.16 |
TRIM7 |
tripartite motif containing 7 |
153 |
0.85 |
| chr15_92400679_92401101 | 0.16 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
3539 |
0.31 |
| chr8_71128828_71128979 | 0.16 |
NCOA2 |
nuclear receptor coactivator 2 |
28707 |
0.22 |
| chr21_47789248_47789399 | 0.16 |
PCNT |
pericentrin |
45287 |
0.1 |
| chr2_220251514_220252585 | 0.16 |
DNPEP |
aspartyl aminopeptidase |
19 |
0.94 |
| chr19_54783961_54784112 | 0.16 |
LILRB2 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
317 |
0.74 |
| chr19_38494188_38494741 | 0.16 |
CTC-244M17.1 |
|
39971 |
0.13 |
| chr2_31541639_31542033 | 0.16 |
EHD3 |
EH-domain containing 3 |
84633 |
0.08 |
| chr6_1624554_1625514 | 0.16 |
FOXC1 |
forkhead box C1 |
14353 |
0.29 |
| chrX_68384708_68385863 | 0.16 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
5 |
0.99 |
| chrX_12929174_12929325 | 0.16 |
TLR8-AS1 |
TLR8 antisense RNA 1 |
2797 |
0.26 |
| chr3_101797946_101798097 | 0.16 |
ZPLD1 |
zona pellucida-like domain containing 1 |
20067 |
0.26 |
| chr15_50356488_50356639 | 0.16 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
16979 |
0.23 |
| chr12_92755876_92756027 | 0.16 |
RP11-693J15.4 |
|
59356 |
0.12 |
| chr10_14920855_14921866 | 0.16 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
439 |
0.82 |
| chr1_167572076_167572227 | 0.16 |
RCSD1 |
RCSD domain containing 1 |
27179 |
0.16 |
| chr2_113885287_113885446 | 0.16 |
IL1RN |
interleukin 1 receptor antagonist |
228 |
0.91 |
| chr11_67807279_67808325 | 0.16 |
TCIRG1 |
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 |
1229 |
0.3 |
| chr3_4793449_4793634 | 0.16 |
ENSG00000239126 |
. |
126845 |
0.05 |
| chr3_32344642_32344793 | 0.16 |
ENSG00000207857 |
. |
43497 |
0.14 |
| chr2_43188289_43188440 | 0.16 |
ENSG00000207087 |
. |
130268 |
0.05 |
| chr18_74843262_74844680 | 0.16 |
MBP |
myelin basic protein |
331 |
0.94 |
| chr10_70883973_70885223 | 0.15 |
VPS26A |
vacuolar protein sorting 26 homolog A (S. pombe) |
595 |
0.74 |
| chr2_197076357_197076527 | 0.15 |
ENSG00000239161 |
. |
3604 |
0.24 |
| chr15_83540435_83540586 | 0.15 |
HOMER2 |
homer homolog 2 (Drosophila) |
7499 |
0.17 |
| chr19_2494417_2495562 | 0.15 |
ENSG00000252962 |
. |
8150 |
0.15 |
| chr2_217180593_217180957 | 0.15 |
AC069155.1 |
|
12727 |
0.18 |
| chr3_15578534_15578685 | 0.15 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
15351 |
0.14 |
| chr9_117568588_117568807 | 0.15 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
291 |
0.92 |
| chr9_124016304_124016710 | 0.15 |
GSN |
gelsolin |
13831 |
0.14 |
| chr9_33523659_33524999 | 0.15 |
ANKRD18B |
ankyrin repeat domain 18B |
63 |
0.97 |
| chr19_45754620_45755932 | 0.15 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
726 |
0.57 |
| chr14_21249278_21249626 | 0.15 |
RNASE6 |
ribonuclease, RNase A family, k6 |
242 |
0.86 |
| chrX_13751428_13751579 | 0.15 |
TRAPPC2 |
trafficking protein particle complex 2 |
1172 |
0.37 |
| chr5_10607962_10608181 | 0.15 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
20266 |
0.2 |
| chr16_1991808_1991959 | 0.15 |
MSRB1 |
methionine sulfoxide reductase B1 |
1241 |
0.18 |
| chrX_13955774_13956778 | 0.15 |
GPM6B |
glycoprotein M6B |
258 |
0.95 |
| chr2_28811248_28811399 | 0.15 |
PLB1 |
phospholipase B1 |
13463 |
0.23 |
| chr6_156338794_156338945 | 0.15 |
ENSG00000221456 |
. |
70938 |
0.14 |
| chr12_48212353_48213256 | 0.15 |
HDAC7 |
histone deacetylase 7 |
241 |
0.91 |
| chr12_92424206_92424357 | 0.15 |
C12orf79 |
chromosome 12 open reading frame 79 |
106516 |
0.07 |
| chr2_190444828_190446170 | 0.15 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
114 |
0.98 |
| chr5_142193879_142194030 | 0.15 |
ARHGAP26 |
Rho GTPase activating protein 26 |
41672 |
0.17 |
| chr18_53006641_53006980 | 0.15 |
TCF4 |
transcription factor 4 |
12418 |
0.28 |
| chr8_142105945_142106235 | 0.15 |
DENND3 |
DENN/MADD domain containing 3 |
21287 |
0.19 |
| chr2_238395971_238396408 | 0.15 |
MLPH |
melanophilin |
270 |
0.92 |
| chr22_31631160_31631361 | 0.15 |
ENSG00000202019 |
. |
5102 |
0.12 |
| chr2_108443697_108444171 | 0.15 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
541 |
0.87 |
| chr2_155001115_155001266 | 0.15 |
ENSG00000266512 |
. |
1530 |
0.54 |
| chr22_17598033_17598230 | 0.15 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
4012 |
0.17 |
| chr2_60774113_60774264 | 0.15 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
6352 |
0.26 |
| chr1_16769456_16769607 | 0.15 |
NECAP2 |
NECAP endocytosis associated 2 |
2300 |
0.23 |
| chr11_126188773_126189028 | 0.15 |
RP11-712L6.5 |
Uncharacterized protein |
14687 |
0.1 |
| chr1_45274565_45275349 | 0.15 |
BTBD19 |
BTB (POZ) domain containing 19 |
762 |
0.37 |
| chr20_48301152_48301303 | 0.15 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
29188 |
0.16 |
| chr1_234658507_234658658 | 0.15 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
43733 |
0.14 |
| chr12_11926562_11926926 | 0.15 |
ETV6 |
ets variant 6 |
21309 |
0.26 |
| chr12_121340609_121341729 | 0.15 |
SPPL3 |
signal peptide peptidase like 3 |
4 |
0.98 |
| chr4_26263054_26263205 | 0.15 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
11100 |
0.31 |
| chr7_150131124_150131275 | 0.15 |
GIMAP8 |
GTPase, IMAP family member 8 |
16519 |
0.14 |
| chr7_105318075_105318226 | 0.15 |
ATXN7L1 |
ataxin 7-like 1 |
1459 |
0.53 |
| chr15_50359492_50359643 | 0.15 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
19983 |
0.22 |
| chr9_5493967_5494181 | 0.15 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
16471 |
0.2 |
| chr2_208091726_208091877 | 0.15 |
AC007879.5 |
|
27175 |
0.18 |
| chr2_30454378_30455881 | 0.14 |
LBH |
limb bud and heart development |
83 |
0.98 |
| chr3_51691608_51691759 | 0.14 |
TEX264 |
testis expressed 264 |
5026 |
0.17 |
| chr13_88325876_88326239 | 0.14 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
559 |
0.85 |
| chr16_29471210_29471476 | 0.14 |
SULT1A4 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
133 |
0.91 |
| chr12_117536177_117536841 | 0.14 |
TESC |
tescalcin |
742 |
0.76 |
| chr19_14551433_14551660 | 0.14 |
PKN1 |
protein kinase N1 |
474 |
0.7 |
| chr16_53124890_53125176 | 0.14 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
8039 |
0.24 |
| chr8_68296421_68296572 | 0.14 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
40584 |
0.21 |
| chr3_131753321_131753947 | 0.14 |
CPNE4 |
copine IV |
210 |
0.96 |
| chr17_40811022_40812282 | 0.14 |
TUBG2 |
tubulin, gamma 2 |
329 |
0.76 |
| chr19_38736860_38737011 | 0.14 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
2260 |
0.16 |
| chr8_77320453_77320604 | 0.14 |
ENSG00000222231 |
. |
142561 |
0.05 |
| chr4_11652870_11653079 | 0.14 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
221585 |
0.02 |
| chr11_118023255_118024161 | 0.14 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
105 |
0.96 |
| chr12_132475297_132475448 | 0.14 |
ENSG00000271905 |
. |
10948 |
0.16 |
| chr21_45527300_45528294 | 0.14 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
543 |
0.75 |
| chr20_3189609_3190930 | 0.14 |
ITPA |
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
98 |
0.95 |
| chr1_158218516_158218667 | 0.14 |
CD1A |
CD1a molecule |
5336 |
0.22 |
| chr2_219124080_219124231 | 0.14 |
GPBAR1 |
G protein-coupled bile acid receptor 1 |
64 |
0.94 |
| chr5_63461313_63462499 | 0.14 |
RNF180 |
ring finger protein 180 |
83 |
0.99 |
| chr15_48604529_48604680 | 0.14 |
ENSG00000202542 |
. |
13264 |
0.17 |
| chr2_198167617_198167768 | 0.14 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
449 |
0.8 |
| chr17_76879212_76879961 | 0.14 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
9354 |
0.14 |
| chr22_30053890_30054041 | 0.14 |
NF2 |
neurofibromin 2 (merlin) |
53977 |
0.08 |
| chr1_207244470_207244690 | 0.14 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
6231 |
0.14 |
| chr7_148844568_148845756 | 0.14 |
ZNF398 |
zinc finger protein 398 |
57 |
0.97 |
| chr17_2699734_2700214 | 0.14 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
198 |
0.94 |
| chr1_170633420_170634117 | 0.14 |
PRRX1 |
paired related homeobox 1 |
690 |
0.8 |
| chr10_22894404_22894761 | 0.14 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
13940 |
0.29 |
| chr4_38802207_38802358 | 0.14 |
TLR1 |
toll-like receptor 1 |
3715 |
0.19 |
| chr11_75490080_75490231 | 0.14 |
CTD-2530H12.2 |
|
3206 |
0.13 |
| chr6_88875424_88876640 | 0.14 |
CNR1 |
cannabinoid receptor 1 (brain) |
46 |
0.99 |
| chr14_89290449_89291591 | 0.14 |
TTC8 |
tetratricopeptide repeat domain 8 |
9 |
0.97 |
| chr10_99090099_99090250 | 0.14 |
RP11-452K12.4 |
|
4147 |
0.13 |
| chr7_31851375_31851526 | 0.14 |
ENSG00000223070 |
. |
17588 |
0.29 |
| chr5_15704054_15704205 | 0.14 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
88038 |
0.09 |
| chr6_53109316_53109571 | 0.14 |
ENSG00000206908 |
. |
25384 |
0.15 |
| chr4_113626760_113627887 | 0.14 |
ENSG00000202536 |
. |
49750 |
0.09 |
| chr1_28571461_28571612 | 0.14 |
RP5-1092A3.4 |
|
3572 |
0.14 |
| chr18_53007051_53007202 | 0.14 |
TCF4 |
transcription factor 4 |
12102 |
0.28 |
| chr17_61106096_61106247 | 0.14 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
19250 |
0.23 |
| chr19_36207272_36208543 | 0.14 |
KMT2B |
Histone-lysine N-methyltransferase 2B |
1014 |
0.26 |
| chr9_88137440_88138079 | 0.14 |
AGTPBP1 |
ATP/GTP binding protein 1 |
158442 |
0.04 |
| chr17_77019963_77021139 | 0.14 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
208 |
0.91 |
| chr16_67282110_67283381 | 0.13 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
108 |
0.89 |
| chr7_148237370_148237521 | 0.13 |
C7orf33 |
chromosome 7 open reading frame 33 |
50212 |
0.15 |
| chr11_67041225_67041376 | 0.13 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
7348 |
0.12 |
| chr3_47031255_47031478 | 0.13 |
NBEAL2 |
neurobeachin-like 2 |
5445 |
0.19 |
| chr20_4807814_4807965 | 0.13 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
3598 |
0.24 |
| chr14_102320200_102320351 | 0.13 |
CTD-2017C7.1 |
|
14407 |
0.16 |
| chr20_47414178_47414329 | 0.13 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
30167 |
0.21 |
| chr15_100017896_100018047 | 0.13 |
MEF2A |
myocyte enhancer factor 2A |
601 |
0.77 |
| chr19_47610168_47610319 | 0.13 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
6288 |
0.16 |
| chr10_81026521_81026806 | 0.13 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
39312 |
0.18 |
| chr1_23860213_23860364 | 0.13 |
E2F2 |
E2F transcription factor 2 |
2576 |
0.26 |
| chr11_119953003_119953290 | 0.13 |
TRIM29 |
tripartite motif containing 29 |
38462 |
0.18 |
| chr14_24803093_24804202 | 0.13 |
ADCY4 |
adenylate cyclase 4 |
306 |
0.72 |
| chr8_27262752_27262933 | 0.13 |
PTK2B |
protein tyrosine kinase 2 beta |
7873 |
0.23 |
| chr19_14223228_14223791 | 0.13 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
1491 |
0.22 |
| chr6_20267392_20267543 | 0.13 |
MBOAT1 |
membrane bound O-acyltransferase domain containing 1 |
54797 |
0.13 |
| chr2_11483078_11484442 | 0.13 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
951 |
0.65 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus(GO:0002866) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.2 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.2 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.2 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0030146 | obsolete diuresis(GO:0030146) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.0 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0001787 | natural killer cell proliferation(GO:0001787) regulation of natural killer cell proliferation(GO:0032817) |
| 0.0 | 0.0 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:1901978 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0051318 | G1 phase(GO:0051318) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.0 | 0.0 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.1 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.0 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0033558 | protein deacetylase activity(GO:0033558) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.0 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |