Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
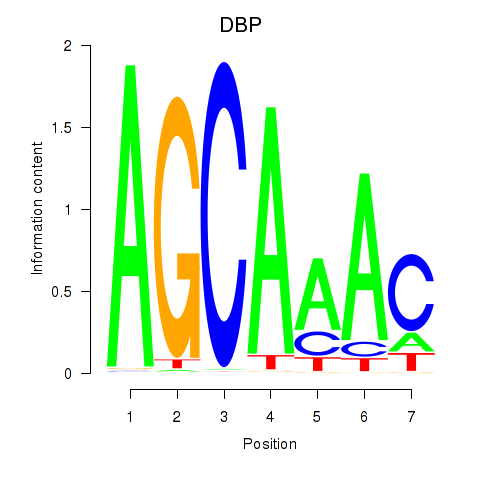
Results for DBP
Z-value: 1.43
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | D-box binding PAR bZIP transcription factor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
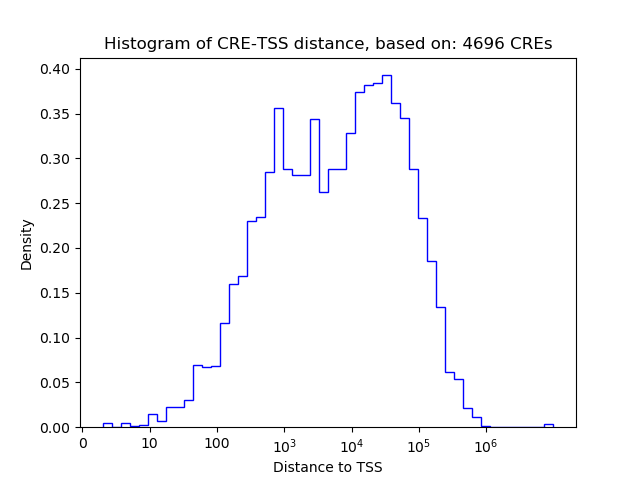
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_49138973_49139430 | DBP | 1308 | 0.220749 | -0.88 | 1.8e-03 | Click! |
| chr19_49139505_49139656 | DBP | 1040 | 0.284260 | -0.84 | 4.8e-03 | Click! |
| chr19_49139809_49140053 | DBP | 689 | 0.437960 | -0.75 | 2.0e-02 | Click! |
| chr19_49140164_49140638 | DBP | 219 | 0.831467 | -0.70 | 3.4e-02 | Click! |
| chr19_49133396_49133547 | DBP | 4312 | 0.088905 | 0.70 | 3.6e-02 | Click! |
Activity of the DBP motif across conditions
Conditions sorted by the z-value of the DBP motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
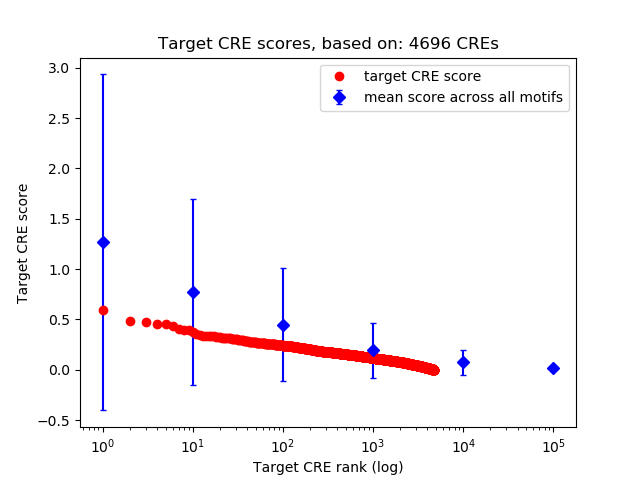
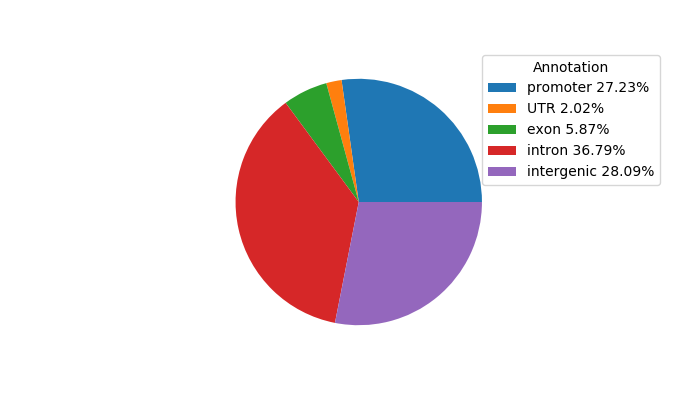
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr21_28214648_28215240 | 0.59 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
393 |
0.9 |
| chrX_30327007_30327420 | 0.49 |
NR0B1 |
nuclear receptor subfamily 0, group B, member 1 |
282 |
0.91 |
| chr3_137486919_137487668 | 0.47 |
SOX14 |
SRY (sex determining region Y)-box 14 |
3714 |
0.37 |
| chr3_149094469_149095370 | 0.46 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
646 |
0.47 |
| chr7_28725956_28726399 | 0.45 |
CREB5 |
cAMP responsive element binding protein 5 |
579 |
0.86 |
| chr1_157980267_157980690 | 0.44 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
14862 |
0.2 |
| chr20_14317720_14318261 | 0.40 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
264 |
0.94 |
| chr13_67708586_67709053 | 0.40 |
PCDH9 |
protocadherin 9 |
93753 |
0.09 |
| chr6_27560724_27561227 | 0.40 |
ENSG00000206671 |
. |
3216 |
0.27 |
| chr9_14400502_14400713 | 0.38 |
NFIB |
nuclear factor I/B |
1625 |
0.51 |
| chr5_54456050_54456451 | 0.36 |
GPX8 |
glutathione peroxidase 8 (putative) |
252 |
0.86 |
| chr5_83017716_83018280 | 0.35 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
566 |
0.78 |
| chr1_99128119_99128537 | 0.34 |
SNX7 |
sorting nexin 7 |
549 |
0.88 |
| chr20_11871763_11872529 | 0.34 |
BTBD3 |
BTB (POZ) domain containing 3 |
171 |
0.97 |
| chr5_73110322_73110752 | 0.34 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
1194 |
0.51 |
| chr6_19838575_19839711 | 0.34 |
RP1-167F1.2 |
|
168 |
0.95 |
| chr13_93879365_93880233 | 0.33 |
GPC6 |
glypican 6 |
704 |
0.82 |
| chr7_22538999_22539909 | 0.33 |
STEAP1B |
STEAP family member 1B |
342 |
0.91 |
| chr6_1821527_1821700 | 0.32 |
FOXC1 |
forkhead box C1 |
210932 |
0.02 |
| chr15_39873424_39874712 | 0.32 |
THBS1 |
thrombospondin 1 |
774 |
0.66 |
| chr2_164592268_164592669 | 0.32 |
FIGN |
fidgetin |
49 |
0.99 |
| chr7_94025115_94025296 | 0.32 |
COL1A2 |
collagen, type I, alpha 2 |
1332 |
0.57 |
| chr2_106260790_106261225 | 0.32 |
NCK2 |
NCK adaptor protein 2 |
100347 |
0.08 |
| chr8_23711881_23712175 | 0.32 |
STC1 |
stanniocalcin 1 |
204 |
0.95 |
| chr4_111552866_111553567 | 0.31 |
PITX2 |
paired-like homeodomain 2 |
4960 |
0.32 |
| chr12_21652771_21652922 | 0.31 |
RECQL |
RecQ protein-like (DNA helicase Q1-like) |
1639 |
0.29 |
| chr1_157979860_157980080 | 0.31 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
15370 |
0.2 |
| chr3_64253305_64253544 | 0.31 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
231 |
0.95 |
| chr12_53443889_53444561 | 0.31 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
255 |
0.86 |
| chr2_238322401_238322657 | 0.31 |
COL6A3 |
collagen, type VI, alpha 3 |
262 |
0.93 |
| chr15_96869491_96869741 | 0.30 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
449 |
0.57 |
| chr5_38846105_38846894 | 0.30 |
OSMR |
oncostatin M receptor |
398 |
0.91 |
| chr9_18476160_18476529 | 0.30 |
ADAMTSL1 |
ADAMTS-like 1 |
2113 |
0.45 |
| chr18_72509166_72509431 | 0.29 |
ZNF407 |
zinc finger protein 407 |
166322 |
0.04 |
| chr6_128828944_128829440 | 0.29 |
RP1-86D1.4 |
|
2922 |
0.22 |
| chr7_54972295_54972520 | 0.29 |
ENSG00000252054 |
. |
38896 |
0.2 |
| chr13_38442983_38443134 | 0.29 |
TRPC4 |
transient receptor potential cation channel, subfamily C, member 4 |
802 |
0.72 |
| chr16_86603613_86603777 | 0.29 |
RP11-463O9.5 |
|
2328 |
0.23 |
| chr1_157964729_157965124 | 0.29 |
KIRREL |
kin of IRRE like (Drosophila) |
1491 |
0.45 |
| chr2_216296824_216297216 | 0.29 |
FN1 |
fibronectin 1 |
3770 |
0.26 |
| chr17_29887718_29887925 | 0.28 |
ENSG00000207614 |
. |
806 |
0.46 |
| chr22_28181066_28181439 | 0.28 |
MN1 |
meningioma (disrupted in balanced translocation) 1 |
16234 |
0.25 |
| chr2_46165643_46166035 | 0.28 |
PRKCE |
protein kinase C, epsilon |
62202 |
0.14 |
| chr3_50395480_50396917 | 0.28 |
TMEM115 |
transmembrane protein 115 |
843 |
0.33 |
| chr6_7697968_7698508 | 0.28 |
BMP6 |
bone morphogenetic protein 6 |
28792 |
0.23 |
| chr12_120241388_120241613 | 0.28 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
313 |
0.92 |
| chr11_8830922_8831183 | 0.27 |
ST5 |
suppression of tumorigenicity 5 |
1144 |
0.43 |
| chr2_119532290_119532494 | 0.27 |
EN1 |
engrailed homeobox 1 |
72862 |
0.12 |
| chr13_33543979_33544184 | 0.27 |
KL |
klotho |
46126 |
0.17 |
| chr9_118918844_118919316 | 0.27 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
2997 |
0.34 |
| chr4_124339787_124340099 | 0.27 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
18820 |
0.3 |
| chr19_4575630_4575781 | 0.27 |
SEMA6B |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
15885 |
0.1 |
| chr1_201915320_201915636 | 0.27 |
LMOD1 |
leiomodin 1 (smooth muscle) |
237 |
0.87 |
| chr6_90927639_90927889 | 0.27 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
78697 |
0.1 |
| chr7_139899946_139900413 | 0.27 |
ENSG00000199971 |
. |
9292 |
0.16 |
| chr15_100651358_100651509 | 0.27 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
2121 |
0.32 |
| chr18_71356617_71356768 | 0.27 |
ENSG00000241866 |
. |
55478 |
0.18 |
| chr2_119603311_119603908 | 0.27 |
EN1 |
engrailed homeobox 1 |
1645 |
0.51 |
| chr4_188523294_188523560 | 0.27 |
ZFP42 |
ZFP42 zinc finger protein |
393498 |
0.01 |
| chr9_79520083_79520617 | 0.26 |
PRUNE2 |
prune homolog 2 (Drosophila) |
651 |
0.82 |
| chr5_72746721_72747193 | 0.26 |
FOXD1 |
forkhead box D1 |
2605 |
0.29 |
| chr1_214397865_214398571 | 0.26 |
SMYD2 |
SET and MYND domain containing 2 |
56358 |
0.16 |
| chr4_169553568_169553728 | 0.26 |
PALLD |
palladin, cytoskeletal associated protein |
880 |
0.63 |
| chr17_13505362_13505866 | 0.26 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
370 |
0.91 |
| chr18_57326676_57327097 | 0.26 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
6508 |
0.21 |
| chr15_59663421_59663716 | 0.26 |
FAM81A |
family with sequence similarity 81, member A |
1324 |
0.28 |
| chr3_114342653_114342865 | 0.26 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
294 |
0.95 |
| chr4_170190156_170190307 | 0.26 |
SH3RF1 |
SH3 domain containing ring finger 1 |
877 |
0.72 |
| chr9_91793130_91793737 | 0.26 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
249 |
0.95 |
| chr10_104536223_104536534 | 0.26 |
WBP1L |
WW domain binding protein 1-like |
372 |
0.83 |
| chr20_57089963_57090433 | 0.26 |
APCDD1L |
adenomatosis polyposis coli down-regulated 1-like |
11 |
0.98 |
| chr3_112354765_112355126 | 0.26 |
CCDC80 |
coiled-coil domain containing 80 |
1999 |
0.4 |
| chr8_93113958_93114847 | 0.26 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
1052 |
0.69 |
| chr1_98514742_98514981 | 0.26 |
ENSG00000225206 |
. |
3134 |
0.4 |
| chr4_81192543_81192855 | 0.26 |
FGF5 |
fibroblast growth factor 5 |
4906 |
0.28 |
| chr4_81392876_81393298 | 0.26 |
C4orf22 |
chromosome 4 open reading frame 22 |
136164 |
0.05 |
| chr3_112929944_112930573 | 0.26 |
BOC |
BOC cell adhesion associated, oncogene regulated |
71 |
0.98 |
| chrX_102940107_102940387 | 0.25 |
MORF4L2 |
mortality factor 4 like 2 |
902 |
0.43 |
| chr2_238321225_238321570 | 0.25 |
COL6A3 |
collagen, type VI, alpha 3 |
1394 |
0.45 |
| chr15_63673315_63673584 | 0.25 |
CA12 |
carbonic anhydrase XII |
585 |
0.82 |
| chr6_139116835_139117557 | 0.25 |
ECT2L |
epithelial cell transforming sequence 2 oncogene-like |
61 |
0.98 |
| chr4_7873664_7873998 | 0.25 |
AFAP1 |
actin filament associated protein 1 |
22 |
0.98 |
| chr2_45396710_45397149 | 0.25 |
SIX2 |
SIX homeobox 2 |
160360 |
0.04 |
| chr22_46472803_46473256 | 0.25 |
FLJ27365 |
hsa-mir-4763 |
3163 |
0.13 |
| chr19_16179581_16179939 | 0.25 |
TPM4 |
tropomyosin 4 |
1250 |
0.44 |
| chr9_112543764_112544054 | 0.25 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
1140 |
0.28 |
| chr7_5634551_5634949 | 0.25 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
433 |
0.81 |
| chr6_28175291_28175856 | 0.25 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
17091 |
0.12 |
| chr15_51386054_51386865 | 0.25 |
TNFAIP8L3 |
tumor necrosis factor, alpha-induced protein 8-like 3 |
11014 |
0.22 |
| chr11_19367315_19367499 | 0.25 |
NAV2 |
neuron navigator 2 |
4864 |
0.23 |
| chr2_47269894_47270523 | 0.25 |
AC093732.1 |
|
2545 |
0.28 |
| chr7_80548112_80548336 | 0.25 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
275 |
0.96 |
| chr6_27205685_27206711 | 0.24 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr7_55087572_55088165 | 0.24 |
EGFR |
epidermal growth factor receptor |
1057 |
0.68 |
| chr3_124604600_124604823 | 0.24 |
ITGB5 |
integrin, beta 5 |
1433 |
0.47 |
| chr14_53416235_53416562 | 0.24 |
FERMT2 |
fermitin family member 2 |
888 |
0.69 |
| chr3_15837119_15837337 | 0.24 |
ANKRD28 |
ankyrin repeat domain 28 |
800 |
0.67 |
| chr3_99357431_99357620 | 0.24 |
COL8A1 |
collagen, type VIII, alpha 1 |
71 |
0.98 |
| chr14_30613029_30613261 | 0.24 |
PRKD1 |
protein kinase D1 |
47959 |
0.19 |
| chr17_881575_881780 | 0.24 |
NXN |
nucleoredoxin |
1333 |
0.34 |
| chr14_61104683_61105223 | 0.24 |
SIX1 |
SIX homeobox 1 |
11227 |
0.21 |
| chr1_157013915_157014350 | 0.24 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
733 |
0.68 |
| chr9_109626964_109627115 | 0.24 |
ZNF462 |
zinc finger protein 462 |
1592 |
0.44 |
| chr2_198539523_198539712 | 0.24 |
RFTN2 |
raftlin family member 2 |
1102 |
0.51 |
| chr2_66664178_66664750 | 0.24 |
MEIS1 |
Meis homeobox 1 |
61 |
0.96 |
| chr8_120685506_120685944 | 0.24 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
32 |
0.98 |
| chr8_18744322_18744566 | 0.24 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
100 |
0.98 |
| chr12_115120954_115121265 | 0.24 |
TBX3 |
T-box 3 |
286 |
0.93 |
| chr1_240855293_240855444 | 0.24 |
ENSG00000199241 |
. |
6252 |
0.23 |
| chr8_49833429_49833759 | 0.24 |
SNAI2 |
snail family zinc finger 2 |
394 |
0.92 |
| chr6_85823864_85824548 | 0.24 |
NT5E |
5'-nucleotidase, ecto (CD73) |
335603 |
0.01 |
| chr5_83679357_83679570 | 0.24 |
EDIL3 |
EGF-like repeats and discoidin I-like domains 3 |
741 |
0.46 |
| chr9_113799034_113800104 | 0.24 |
LPAR1 |
lysophosphatidic acid receptor 1 |
754 |
0.74 |
| chr11_120208718_120208869 | 0.24 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
847 |
0.6 |
| chr15_30112674_30113382 | 0.24 |
TJP1 |
tight junction protein 1 |
723 |
0.64 |
| chr2_9348603_9349207 | 0.24 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
2011 |
0.46 |
| chr1_68084153_68084510 | 0.23 |
ENSG00000242482 |
. |
38481 |
0.16 |
| chr4_81188224_81188744 | 0.23 |
FGF5 |
fibroblast growth factor 5 |
691 |
0.76 |
| chr12_125169284_125169605 | 0.23 |
NCOR2 |
nuclear receptor corepressor 2 |
117434 |
0.06 |
| chr3_141087883_141088209 | 0.23 |
RP11-438D8.2 |
|
2067 |
0.36 |
| chr21_45006172_45006433 | 0.23 |
HSF2BP |
heat shock transcription factor 2 binding protein |
71723 |
0.1 |
| chr4_129307789_129307940 | 0.23 |
PGRMC2 |
progesterone receptor membrane component 2 |
97880 |
0.09 |
| chr3_25469138_25469713 | 0.23 |
RARB |
retinoic acid receptor, beta |
329 |
0.93 |
| chr3_138892567_138893067 | 0.23 |
MRPS22 |
mitochondrial ribosomal protein S22 |
63571 |
0.12 |
| chr4_126311427_126311944 | 0.23 |
FAT4 |
FAT atypical cadherin 4 |
3406 |
0.33 |
| chr3_129407054_129407730 | 0.23 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
183 |
0.96 |
| chr1_185703938_185704134 | 0.23 |
HMCN1 |
hemicentin 1 |
353 |
0.92 |
| chr12_47219516_47219750 | 0.23 |
SLC38A4 |
solute carrier family 38, member 4 |
135 |
0.98 |
| chr5_82768204_82768925 | 0.23 |
VCAN |
versican |
820 |
0.75 |
| chr12_72665374_72665945 | 0.23 |
ENSG00000236333 |
. |
112 |
0.93 |
| chr5_162992519_162993506 | 0.23 |
MAT2B |
methionine adenosyltransferase II, beta |
60392 |
0.11 |
| chr15_80988981_80989242 | 0.23 |
ABHD17C |
abhydrolase domain containing 17C |
1459 |
0.44 |
| chr2_133998643_133998815 | 0.23 |
AC010890.1 |
|
23811 |
0.24 |
| chr3_159484310_159484461 | 0.23 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1493 |
0.34 |
| chr11_79151385_79151576 | 0.23 |
TENM4 |
teneurin transmembrane protein 4 |
215 |
0.95 |
| chr4_71996832_71997002 | 0.23 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
56086 |
0.15 |
| chr8_18870397_18870669 | 0.23 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
663 |
0.74 |
| chr7_41735925_41736162 | 0.23 |
INHBA-AS1 |
INHBA antisense RNA 1 |
2497 |
0.29 |
| chr6_16200733_16201077 | 0.22 |
ENSG00000251831 |
. |
4340 |
0.21 |
| chr6_85473331_85473901 | 0.22 |
TBX18 |
T-box 18 |
445 |
0.9 |
| chr5_78985270_78986298 | 0.22 |
CMYA5 |
cardiomyopathy associated 5 |
84 |
0.98 |
| chr18_55889348_55889572 | 0.22 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
657 |
0.77 |
| chr5_111091378_111091829 | 0.22 |
NREP |
neuronal regeneration related protein |
345 |
0.9 |
| chr18_57363476_57363657 | 0.22 |
RP11-2N1.2 |
|
125 |
0.93 |
| chr7_132737872_132738088 | 0.22 |
CHCHD3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
28820 |
0.2 |
| chr18_56244152_56244311 | 0.22 |
RP11-126O1.2 |
|
19929 |
0.14 |
| chr8_13132829_13133895 | 0.22 |
DLC1 |
deleted in liver cancer 1 |
693 |
0.78 |
| chr1_67000065_67000303 | 0.22 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
219 |
0.96 |
| chr15_96880233_96880461 | 0.22 |
ENSG00000222651 |
. |
3857 |
0.18 |
| chrX_38079311_38080259 | 0.22 |
SRPX |
sushi-repeat containing protein, X-linked |
367 |
0.75 |
| chr10_6214045_6214449 | 0.22 |
ENSG00000263628 |
. |
20088 |
0.13 |
| chr12_66220556_66220804 | 0.22 |
HMGA2 |
high mobility group AT-hook 2 |
1777 |
0.39 |
| chr5_37839467_37839691 | 0.22 |
GDNF |
glial cell derived neurotrophic factor |
203 |
0.96 |
| chr7_114055343_114055580 | 0.22 |
FOXP2 |
forkhead box P2 |
83 |
0.98 |
| chr6_10426220_10426432 | 0.22 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
6455 |
0.18 |
| chr11_27743805_27744092 | 0.22 |
BDNF |
brain-derived neurotrophic factor |
343 |
0.92 |
| chrX_107178963_107179207 | 0.22 |
RP6-191P20.4 |
|
125 |
0.97 |
| chr1_13910269_13910760 | 0.22 |
PDPN |
podoplanin |
74 |
0.97 |
| chr3_64671738_64672057 | 0.22 |
ADAMTS9-AS2 |
ADAMTS9 antisense RNA 2 |
1093 |
0.46 |
| chr13_33858654_33859881 | 0.22 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
625 |
0.76 |
| chr10_24755480_24755818 | 0.22 |
KIAA1217 |
KIAA1217 |
189 |
0.96 |
| chr14_42074269_42074448 | 0.22 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
2415 |
0.37 |
| chr21_17791847_17792197 | 0.22 |
ENSG00000207638 |
. |
119387 |
0.06 |
| chr20_35169899_35170178 | 0.21 |
MYL9 |
myosin, light chain 9, regulatory |
138 |
0.95 |
| chr5_87988240_87988451 | 0.21 |
CTC-467M3.2 |
|
123 |
0.97 |
| chr6_11229774_11229925 | 0.21 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
3042 |
0.32 |
| chr3_180397078_180397946 | 0.21 |
CCDC39 |
coiled-coil domain containing 39 |
224 |
0.65 |
| chr18_60153182_60153479 | 0.21 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
36910 |
0.19 |
| chr7_97952929_97953353 | 0.21 |
RP11-307C18.1 |
|
976 |
0.55 |
| chr20_9049623_9049963 | 0.21 |
PLCB4 |
phospholipase C, beta 4 |
45 |
0.98 |
| chr1_101186979_101187209 | 0.21 |
VCAM1 |
vascular cell adhesion molecule 1 |
1774 |
0.47 |
| chr11_121969590_121969741 | 0.21 |
ENSG00000207971 |
. |
887 |
0.47 |
| chr11_122044065_122044553 | 0.21 |
ENSG00000207994 |
. |
21293 |
0.17 |
| chr9_94065254_94065575 | 0.21 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
58763 |
0.16 |
| chr18_51736037_51736198 | 0.21 |
ENSG00000207233 |
. |
12665 |
0.21 |
| chr13_21287159_21287369 | 0.21 |
ENSG00000265710 |
. |
9422 |
0.2 |
| chr2_188418224_188418410 | 0.21 |
AC007319.1 |
|
559 |
0.57 |
| chr20_19738276_19738696 | 0.21 |
AL121761.2 |
Uncharacterized protein |
193 |
0.95 |
| chr6_52530050_52530201 | 0.21 |
RP1-152L7.5 |
|
53 |
0.98 |
| chr6_131291045_131291463 | 0.21 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
369 |
0.93 |
| chr2_167231853_167232143 | 0.21 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
499 |
0.84 |
| chr16_73090314_73090763 | 0.21 |
ZFHX3 |
zinc finger homeobox 3 |
3059 |
0.3 |
| chr9_35945444_35945832 | 0.21 |
OR2S2 |
olfactory receptor, family 2, subfamily S, member 2 |
12513 |
0.14 |
| chr8_32407136_32407287 | 0.21 |
NRG1 |
neuregulin 1 |
966 |
0.71 |
| chr2_158114922_158115143 | 0.21 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
922 |
0.66 |
| chr6_138427325_138427625 | 0.21 |
PERP |
PERP, TP53 apoptosis effector |
1173 |
0.59 |
| chr3_16219029_16219297 | 0.21 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
2947 |
0.33 |
| chr8_12989314_12989523 | 0.21 |
DLC1 |
deleted in liver cancer 1 |
1565 |
0.46 |
| chr9_93644008_93644159 | 0.21 |
SYK |
spleen tyrosine kinase |
54313 |
0.18 |
| chr10_33630504_33630927 | 0.21 |
NRP1 |
neuropilin 1 |
5525 |
0.29 |
| chr6_114180362_114180910 | 0.21 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
2095 |
0.32 |
| chr8_141625870_141626141 | 0.21 |
AGO2 |
argonaute RISC catalytic component 2 |
10018 |
0.24 |
| chr1_227503732_227504041 | 0.21 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
997 |
0.69 |
| chr13_33924410_33924807 | 0.21 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
153 |
0.97 |
| chr6_114663085_114663750 | 0.21 |
HS3ST5 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
792 |
0.72 |
| chr1_20915430_20915891 | 0.21 |
CDA |
cytidine deaminase |
219 |
0.93 |
| chr15_90205872_90206267 | 0.20 |
PLIN1 |
perilipin 1 |
4802 |
0.14 |
| chr2_138722074_138722590 | 0.20 |
HNMT |
histamine N-methyltransferase |
289 |
0.95 |
| chr7_89783827_89784376 | 0.20 |
STEAP1 |
six transmembrane epithelial antigen of the prostate 1 |
412 |
0.81 |
| chr11_79149207_79149581 | 0.20 |
TENM4 |
teneurin transmembrane protein 4 |
2301 |
0.34 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.4 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.1 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.6 | GO:0001946 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.1 | 0.7 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.1 | 0.3 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.3 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.4 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.3 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.2 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.1 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.1 | 0.2 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.2 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.2 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.4 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.1 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.2 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.1 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.2 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.0 | GO:0001840 | neural plate morphogenesis(GO:0001839) neural plate development(GO:0001840) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0043266 | regulation of potassium ion transport(GO:0043266) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.2 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.3 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0030146 | obsolete diuresis(GO:0030146) |
| 0.0 | 0.1 | GO:1903428 | positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0010875 | positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0060405 | regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0072077 | mesenchymal to epithelial transition(GO:0060231) renal vesicle morphogenesis(GO:0072077) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0036126 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 2.1 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 2.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.6 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.3 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0004064 | arylesterase activity(GO:0004064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |