Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
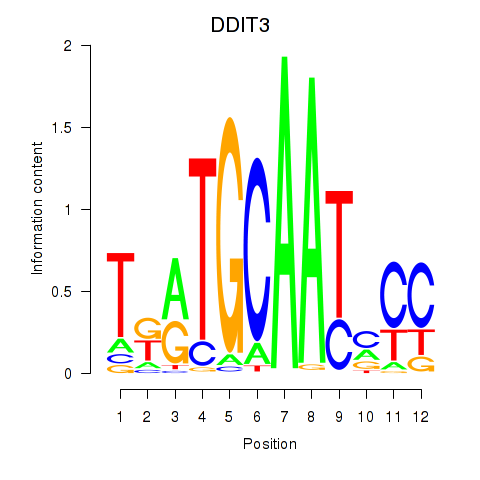
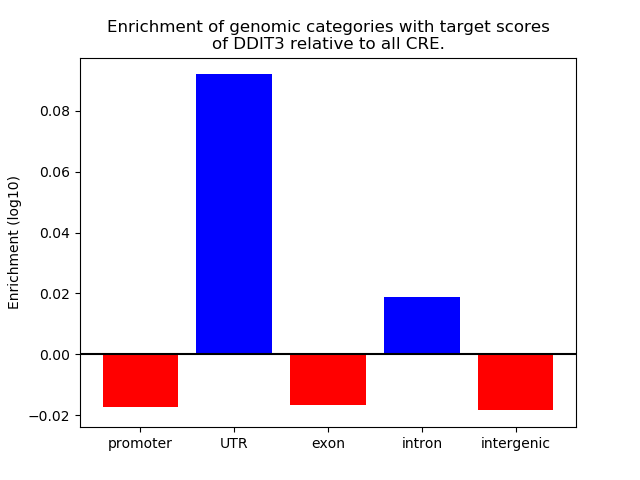
Results for DDIT3
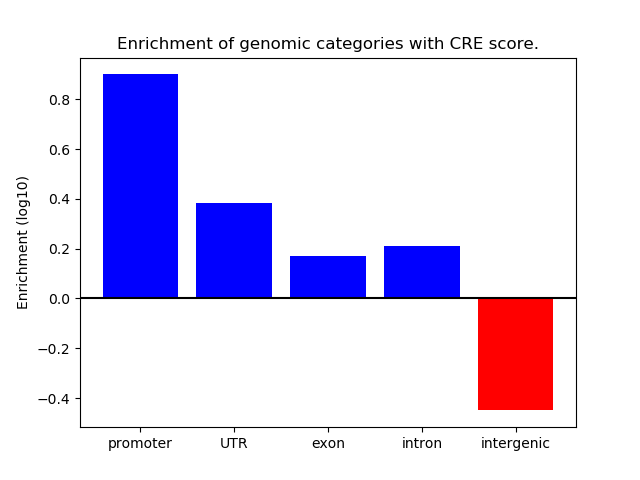
Z-value: 0.90
Transcription factors associated with DDIT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DDIT3
|
ENSG00000175197.6 | DNA damage inducible transcript 3 |
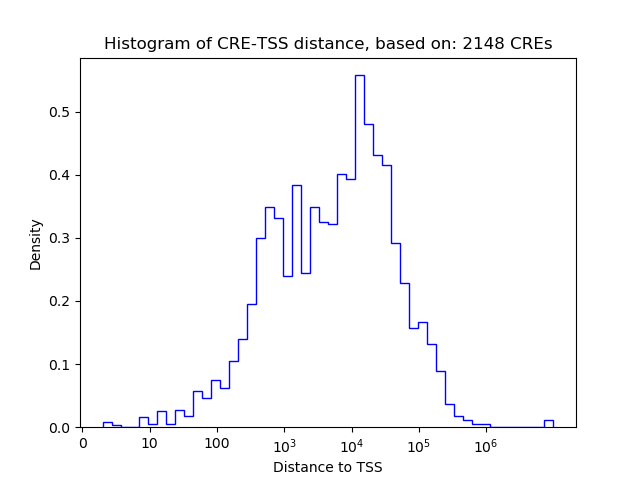
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_57913983_57914218 | DDIT3 | 175 | 0.663482 | -0.23 | 5.5e-01 | Click! |
Activity of the DDIT3 motif across conditions
Conditions sorted by the z-value of the DDIT3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
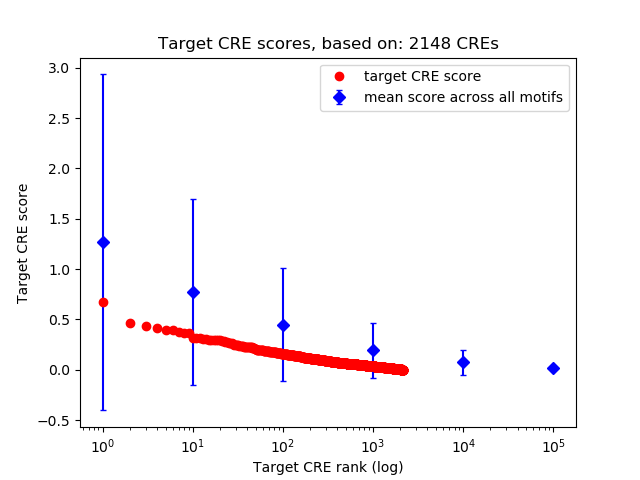
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_36948102_36948314 | 0.68 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
556 |
0.73 |
| chr20_54987227_54988129 | 0.46 |
CASS4 |
Cas scaffolding protein family member 4 |
361 |
0.82 |
| chr13_111314544_111314695 | 0.43 |
CARS2 |
cysteinyl-tRNA synthetase 2, mitochondrial (putative) |
14752 |
0.2 |
| chr13_31312260_31312411 | 0.42 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
2690 |
0.38 |
| chr22_37679801_37680063 | 0.39 |
CYTH4 |
cytohesin 4 |
1404 |
0.39 |
| chr11_121324494_121324645 | 0.39 |
RP11-730K11.1 |
|
847 |
0.59 |
| chr19_16259637_16259788 | 0.37 |
HSH2D |
hematopoietic SH2 domain containing |
5166 |
0.13 |
| chr5_39217983_39218134 | 0.37 |
FYB |
FYN binding protein |
1607 |
0.52 |
| chr2_175532806_175532957 | 0.36 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
14718 |
0.23 |
| chr12_25207113_25207806 | 0.32 |
LRMP |
lymphoid-restricted membrane protein |
1785 |
0.37 |
| chr17_25609205_25609356 | 0.31 |
ENSG00000263583 |
. |
11742 |
0.23 |
| chr22_50523508_50523892 | 0.31 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
12 |
0.97 |
| chr3_196361762_196362043 | 0.31 |
LINC01063 |
long intergenic non-protein coding RNA 1063 |
2444 |
0.19 |
| chr1_111416540_111416879 | 0.31 |
CD53 |
CD53 molecule |
933 |
0.59 |
| chr17_57803272_57803423 | 0.30 |
VMP1 |
vacuole membrane protein 1 |
3729 |
0.21 |
| chr16_57076639_57076877 | 0.30 |
NLRC5 |
NLR family, CARD domain containing 5 |
15219 |
0.13 |
| chr13_46752366_46752713 | 0.29 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
3920 |
0.19 |
| chr8_134151695_134151846 | 0.29 |
TG |
thyroglobulin |
26023 |
0.18 |
| chr5_71768722_71768873 | 0.29 |
RP11-389C8.2 |
|
30583 |
0.19 |
| chr1_17956706_17956857 | 0.29 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
11932 |
0.27 |
| chr1_248020847_248021774 | 0.29 |
TRIM58 |
tripartite motif containing 58 |
809 |
0.49 |
| chr10_11218203_11218399 | 0.28 |
RP3-323N1.2 |
|
4962 |
0.24 |
| chr8_142131426_142131715 | 0.28 |
DENND3 |
DENN/MADD domain containing 3 |
4193 |
0.23 |
| chr21_36280824_36280975 | 0.28 |
RUNX1 |
runt-related transcription factor 1 |
18812 |
0.28 |
| chr6_144655565_144656031 | 0.26 |
RP1-91J24.3 |
|
1361 |
0.51 |
| chr2_111801445_111801596 | 0.26 |
ACOXL |
acyl-CoA oxidase-like |
56745 |
0.14 |
| chr6_24922511_24922662 | 0.26 |
FAM65B |
family with sequence similarity 65, member B |
11391 |
0.23 |
| chr17_45743155_45743306 | 0.26 |
KPNB1 |
karyopherin (importin) beta 1 |
2501 |
0.2 |
| chr1_198612556_198613143 | 0.25 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
4557 |
0.28 |
| chr12_7060265_7060416 | 0.25 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
85 |
0.9 |
| chr2_145340979_145341178 | 0.24 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
62457 |
0.14 |
| chrX_9431281_9431551 | 0.24 |
TBL1X |
transducin (beta)-like 1X-linked |
45 |
0.99 |
| chr15_39745852_39746003 | 0.24 |
RP11-624L4.1 |
|
26531 |
0.23 |
| chr6_34330915_34331066 | 0.24 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
29461 |
0.16 |
| chr19_41303975_41304672 | 0.24 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
578 |
0.64 |
| chr11_12146032_12146187 | 0.23 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
3087 |
0.36 |
| chr5_75697779_75697971 | 0.23 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
1199 |
0.61 |
| chr6_131895116_131895267 | 0.23 |
ARG1 |
arginase 1 |
826 |
0.65 |
| chr10_63819340_63819491 | 0.23 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
10445 |
0.27 |
| chr9_134148708_134148859 | 0.22 |
FAM78A |
family with sequence similarity 78, member A |
2873 |
0.24 |
| chr1_192515169_192515320 | 0.22 |
RP5-1011O1.2 |
|
21103 |
0.19 |
| chr3_99791321_99791472 | 0.22 |
FILIP1L |
filamin A interacting protein 1-like |
41961 |
0.15 |
| chr4_185373022_185373173 | 0.22 |
ENSG00000251878 |
. |
19977 |
0.17 |
| chr10_73520322_73520473 | 0.22 |
C10orf54 |
chromosome 10 open reading frame 54 |
3010 |
0.26 |
| chr20_34356216_34356461 | 0.22 |
ENSG00000222152 |
. |
1451 |
0.3 |
| chr2_197104668_197104938 | 0.22 |
AC020571.3 |
|
19945 |
0.17 |
| chr11_48075840_48075991 | 0.22 |
AC103828.1 |
|
38508 |
0.15 |
| chr15_86098266_86098417 | 0.22 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
336 |
0.89 |
| chr2_161423396_161423547 | 0.20 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
73166 |
0.12 |
| chr19_13206703_13207270 | 0.20 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
6695 |
0.11 |
| chr17_40423913_40424064 | 0.20 |
AC003104.1 |
|
599 |
0.6 |
| chr2_201934315_201934539 | 0.20 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
1729 |
0.24 |
| chr1_160681036_160681588 | 0.20 |
CD48 |
CD48 molecule |
281 |
0.89 |
| chr8_61821706_61823076 | 0.20 |
RP11-33I11.2 |
|
100226 |
0.08 |
| chr13_33608099_33608250 | 0.20 |
ENSG00000221677 |
. |
7315 |
0.24 |
| chr10_6969023_6969174 | 0.20 |
PRKCQ |
protein kinase C, theta |
346835 |
0.01 |
| chr18_60939286_60939489 | 0.20 |
RP11-28F1.2 |
|
41928 |
0.13 |
| chr8_11681660_11681811 | 0.19 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
14970 |
0.13 |
| chr20_30198550_30198775 | 0.19 |
ENSG00000264395 |
. |
3673 |
0.14 |
| chr9_134150778_134151758 | 0.19 |
FAM78A |
family with sequence similarity 78, member A |
666 |
0.69 |
| chr6_86353352_86354469 | 0.19 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
400 |
0.85 |
| chr2_143886362_143886549 | 0.19 |
ARHGAP15 |
Rho GTPase activating protein 15 |
428 |
0.89 |
| chr5_172232754_172232905 | 0.19 |
ENSG00000206741 |
. |
25732 |
0.13 |
| chr8_134086469_134086645 | 0.19 |
SLA |
Src-like-adaptor |
13954 |
0.23 |
| chr2_32784349_32784500 | 0.19 |
ENSG00000207653 |
. |
27204 |
0.2 |
| chrX_38665177_38665418 | 0.19 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
2161 |
0.33 |
| chr9_79252333_79252484 | 0.19 |
PRUNE2 |
prune homolog 2 (Drosophila) |
15057 |
0.21 |
| chr6_24937107_24937405 | 0.19 |
FAM65B |
family with sequence similarity 65, member B |
1068 |
0.59 |
| chr5_17445932_17446083 | 0.19 |
ENSG00000201715 |
. |
100282 |
0.08 |
| chr1_180133242_180133393 | 0.18 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
9285 |
0.24 |
| chr2_153469359_153469510 | 0.18 |
FMNL2 |
formin-like 2 |
6658 |
0.31 |
| chr2_46072609_46072760 | 0.18 |
PRKCE |
protein kinase C, epsilon |
155357 |
0.04 |
| chr4_185773586_185773737 | 0.18 |
ENSG00000266698 |
. |
1397 |
0.43 |
| chr2_135004109_135004260 | 0.18 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
7646 |
0.25 |
| chr9_113065636_113065787 | 0.18 |
TXNDC8 |
thioredoxin domain containing 8 (spermatozoa) |
34377 |
0.16 |
| chr2_37844682_37844833 | 0.18 |
AC006369.2 |
|
17478 |
0.22 |
| chr12_10281805_10282201 | 0.18 |
CLEC7A |
C-type lectin domain family 7, member A |
678 |
0.6 |
| chr12_11803481_11804350 | 0.18 |
ETV6 |
ets variant 6 |
1127 |
0.62 |
| chr20_50267564_50267715 | 0.18 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
88269 |
0.09 |
| chr10_76092336_76092487 | 0.18 |
ADK |
adenosine kinase |
155890 |
0.04 |
| chr17_68166405_68166607 | 0.18 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
830 |
0.45 |
| chr1_40629914_40630065 | 0.17 |
RLF |
rearranged L-myc fusion |
2944 |
0.23 |
| chr18_13464392_13464543 | 0.17 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
547 |
0.62 |
| chr12_32113291_32114105 | 0.17 |
KIAA1551 |
KIAA1551 |
1345 |
0.51 |
| chr22_40858929_40859404 | 0.17 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
256 |
0.92 |
| chr1_246870680_246870831 | 0.17 |
SCCPDH |
saccharopine dehydrogenase (putative) |
16594 |
0.19 |
| chr2_169458585_169458736 | 0.17 |
ENSG00000199348 |
. |
4346 |
0.26 |
| chr16_31207787_31207938 | 0.17 |
RP11-388M20.1 |
|
410 |
0.64 |
| chr1_183549635_183549786 | 0.17 |
NCF2 |
neutrophil cytosolic factor 2 |
10006 |
0.2 |
| chr1_184127977_184128128 | 0.17 |
ENSG00000199840 |
. |
12905 |
0.26 |
| chr5_136949485_136949636 | 0.17 |
ENSG00000216009 |
. |
33778 |
0.18 |
| chr2_8519851_8520002 | 0.16 |
AC011747.7 |
|
295970 |
0.01 |
| chr9_101870409_101870560 | 0.16 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
3052 |
0.29 |
| chr11_65059695_65060120 | 0.16 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
9929 |
0.12 |
| chr11_10480131_10480282 | 0.16 |
AMPD3 |
adenosine monophosphate deaminase 3 |
2473 |
0.29 |
| chr6_159483193_159483344 | 0.16 |
TAGAP |
T-cell activation RhoGTPase activating protein |
17084 |
0.19 |
| chr1_112017238_112017517 | 0.16 |
C1orf162 |
chromosome 1 open reading frame 162 |
886 |
0.41 |
| chr10_21501235_21501386 | 0.16 |
NEBL-AS1 |
NEBL antisense RNA 1 |
38027 |
0.16 |
| chr8_64181080_64181231 | 0.16 |
ENSG00000241964 |
. |
33686 |
0.22 |
| chr18_4862576_4862727 | 0.16 |
ENSG00000222463 |
. |
125517 |
0.06 |
| chr4_177754947_177755098 | 0.16 |
VEGFC |
vascular endothelial growth factor C |
41141 |
0.21 |
| chr20_61200398_61200730 | 0.16 |
ENSG00000207764 |
. |
38445 |
0.09 |
| chr12_15699901_15700074 | 0.16 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
701 |
0.8 |
| chr3_128722868_128723019 | 0.16 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
2471 |
0.22 |
| chr4_12125602_12125753 | 0.16 |
ENSG00000266669 |
. |
110612 |
0.08 |
| chr13_100059124_100059288 | 0.16 |
ENSG00000266207 |
. |
46517 |
0.12 |
| chr17_68316942_68317093 | 0.16 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
151341 |
0.04 |
| chr7_92776252_92776403 | 0.16 |
SAMD9L |
sterile alpha motif domain containing 9-like |
1331 |
0.49 |
| chr21_35917446_35917660 | 0.15 |
RCAN1 |
regulator of calcineurin 1 |
14924 |
0.2 |
| chr10_63818920_63819071 | 0.15 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
10025 |
0.27 |
| chr12_104324193_104325484 | 0.15 |
ENSG00000265072 |
. |
635 |
0.44 |
| chr22_45772663_45772814 | 0.15 |
SMC1B |
structural maintenance of chromosomes 1B |
36710 |
0.13 |
| chr20_10486029_10486187 | 0.15 |
SLX4IP |
SLX4 interacting protein |
70157 |
0.11 |
| chr19_2253059_2253418 | 0.15 |
JSRP1 |
junctional sarcoplasmic reticulum protein 1 |
2110 |
0.13 |
| chr16_87093858_87094180 | 0.15 |
RP11-899L11.3 |
|
155502 |
0.04 |
| chr13_99909010_99909161 | 0.15 |
GPR18 |
G protein-coupled receptor 18 |
1543 |
0.41 |
| chr4_156597025_156597176 | 0.15 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
8285 |
0.26 |
| chr17_71965951_71966102 | 0.15 |
RPL38 |
ribosomal protein L38 |
233695 |
0.02 |
| chr7_38670068_38671048 | 0.15 |
AMPH |
amphiphysin |
462 |
0.87 |
| chr6_14355345_14355496 | 0.15 |
ENSG00000238987 |
. |
198727 |
0.03 |
| chr16_55608580_55608731 | 0.15 |
CAPNS2 |
calpain, small subunit 2 |
8071 |
0.24 |
| chrX_117672480_117672631 | 0.15 |
ENSG00000206862 |
. |
30923 |
0.19 |
| chr12_32742619_32742910 | 0.15 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
8696 |
0.27 |
| chr6_144606475_144607358 | 0.15 |
UTRN |
utrophin |
79 |
0.98 |
| chr2_176399969_176400120 | 0.14 |
ENSG00000221347 |
. |
204943 |
0.02 |
| chr7_7221124_7221275 | 0.14 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
979 |
0.6 |
| chr15_63780195_63780346 | 0.14 |
USP3 |
ubiquitin specific peptidase 3 |
16523 |
0.23 |
| chr10_115604050_115604207 | 0.14 |
DCLRE1A |
DNA cross-link repair 1A |
9731 |
0.17 |
| chr7_150414088_150414577 | 0.14 |
GIMAP1 |
GTPase, IMAP family member 1 |
687 |
0.65 |
| chr1_169580216_169580367 | 0.14 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
8166 |
0.21 |
| chr5_139989047_139989276 | 0.14 |
CD14 |
CD14 molecule |
24075 |
0.07 |
| chr1_208091875_208092026 | 0.14 |
CD34 |
CD34 molecule |
7203 |
0.31 |
| chr19_35837974_35838249 | 0.14 |
ENSG00000263397 |
. |
1695 |
0.19 |
| chr2_113589955_113590106 | 0.14 |
IL1B |
interleukin 1, beta |
3981 |
0.2 |
| chr11_1872722_1872911 | 0.14 |
LSP1 |
lymphocyte-specific protein 1 |
1384 |
0.25 |
| chr14_52313408_52314007 | 0.14 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
137 |
0.97 |
| chr6_12128420_12128571 | 0.14 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
2516 |
0.42 |
| chr3_141121365_141121859 | 0.14 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
48 |
0.98 |
| chr10_20117593_20117744 | 0.14 |
PLXDC2 |
plexin domain containing 2 |
12500 |
0.2 |
| chr15_52844361_52844512 | 0.14 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
5505 |
0.2 |
| chr1_36521425_36521576 | 0.14 |
TEKT2 |
tektin 2 (testicular) |
28176 |
0.13 |
| chr18_21594952_21595765 | 0.14 |
RP11-403A21.2 |
|
55 |
0.94 |
| chr13_53542376_53542527 | 0.14 |
OLFM4 |
olfactomedin 4 |
60443 |
0.14 |
| chr2_134876082_134876233 | 0.14 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
1596 |
0.46 |
| chr22_46988879_46989444 | 0.14 |
GRAMD4 |
GRAM domain containing 4 |
16135 |
0.21 |
| chr1_212781685_212782134 | 0.14 |
ATF3 |
activating transcription factor 3 |
103 |
0.96 |
| chr19_16372792_16372943 | 0.14 |
CTD-2562J15.6 |
|
31519 |
0.12 |
| chr14_22593025_22593176 | 0.14 |
ENSG00000238634 |
. |
17787 |
0.27 |
| chr2_207565040_207565191 | 0.14 |
ENSG00000207319 |
. |
794 |
0.61 |
| chr8_121822100_121822251 | 0.13 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
2208 |
0.3 |
| chr1_160611300_160611550 | 0.13 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
5386 |
0.18 |
| chr6_37770727_37770878 | 0.13 |
RP3-441A12.1 |
|
16192 |
0.18 |
| chr7_130597934_130598170 | 0.13 |
ENSG00000226380 |
. |
35754 |
0.19 |
| chr19_57349494_57349674 | 0.13 |
ZIM2 |
zinc finger, imprinted 2 |
2138 |
0.28 |
| chr13_99964812_99964963 | 0.13 |
GPR183 |
G protein-coupled receptor 183 |
5228 |
0.22 |
| chr1_41827168_41827846 | 0.13 |
FOXO6 |
forkhead box O6 |
87 |
0.98 |
| chr10_3710614_3710914 | 0.13 |
RP11-184A2.3 |
|
82495 |
0.1 |
| chrY_1342944_1343095 | 0.13 |
NA |
NA |
> 106 |
NA |
| chrX_1392949_1393100 | 0.13 |
CSF2RA |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
5286 |
0.11 |
| chr7_26141990_26142141 | 0.13 |
ENSG00000266430 |
. |
42106 |
0.14 |
| chr14_51280333_51280548 | 0.13 |
RP11-286O18.1 |
|
8158 |
0.15 |
| chr7_104581843_104582295 | 0.13 |
ENSG00000251911 |
. |
29665 |
0.15 |
| chr3_113467764_113467962 | 0.13 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
1966 |
0.3 |
| chrX_46509501_46509746 | 0.13 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
76404 |
0.1 |
| chr1_243646030_243646536 | 0.13 |
RP11-269F20.1 |
|
62551 |
0.14 |
| chr10_119165300_119165451 | 0.13 |
PDZD8 |
PDZ domain containing 8 |
30397 |
0.18 |
| chr5_7879366_7879517 | 0.13 |
MTRR |
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
9067 |
0.2 |
| chr5_134695220_134695371 | 0.13 |
C5orf66 |
chromosome 5 open reading frame 66 |
21666 |
0.16 |
| chr13_95133005_95133156 | 0.12 |
DCT |
dopachrome tautomerase |
1144 |
0.56 |
| chr19_16229742_16229926 | 0.12 |
RAB8A |
RAB8A, member RAS oncogene family |
7127 |
0.13 |
| chr16_11208681_11208920 | 0.12 |
CLEC16A |
C-type lectin domain family 16, member A |
8800 |
0.2 |
| chr21_15914846_15914997 | 0.12 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
3741 |
0.3 |
| chr8_37966919_37967070 | 0.12 |
ASH2L |
ash2 (absent, small, or homeotic)-like (Drosophila) |
3537 |
0.14 |
| chr10_11183987_11184138 | 0.12 |
CELF2 |
CUGBP, Elav-like family member 2 |
22931 |
0.18 |
| chr6_155545887_155546038 | 0.12 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
7862 |
0.22 |
| chr12_65032414_65032565 | 0.12 |
RP11-338E21.2 |
|
10365 |
0.12 |
| chr11_2290287_2290803 | 0.12 |
ASCL2 |
achaete-scute family bHLH transcription factor 2 |
1637 |
0.29 |
| chr17_10018080_10018535 | 0.12 |
GAS7 |
growth arrest-specific 7 |
437 |
0.85 |
| chr4_105982764_105983024 | 0.12 |
ENSG00000252136 |
. |
44250 |
0.17 |
| chr3_71626106_71626306 | 0.12 |
FOXP1-IT1 |
FOXP1 intronic transcript 1 (non-protein coding) |
2598 |
0.29 |
| chr6_163826222_163826373 | 0.12 |
QKI |
QKI, KH domain containing, RNA binding |
9378 |
0.32 |
| chr14_65521176_65521327 | 0.12 |
ENSG00000266531 |
. |
9845 |
0.14 |
| chr8_141124879_141125135 | 0.12 |
C8orf17 |
chromosome 8 open reading frame 17 |
181591 |
0.03 |
| chr3_16883275_16883542 | 0.12 |
PLCL2 |
phospholipase C-like 2 |
43044 |
0.19 |
| chr2_30371809_30371960 | 0.12 |
YPEL5 |
yippee-like 5 (Drosophila) |
1502 |
0.4 |
| chr20_52198944_52199193 | 0.12 |
ZNF217 |
zinc finger protein 217 |
568 |
0.75 |
| chr2_208004362_208004513 | 0.12 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
5576 |
0.21 |
| chr4_184580518_184581453 | 0.12 |
TRAPPC11 |
trafficking protein particle complex 11 |
539 |
0.46 |
| chr17_27466417_27467105 | 0.12 |
MYO18A |
myosin XVIIIA |
675 |
0.43 |
| chr1_19642719_19642870 | 0.11 |
ENSG00000265192 |
. |
3760 |
0.13 |
| chr15_64747923_64748074 | 0.11 |
ZNF609 |
zinc finger protein 609 |
4943 |
0.14 |
| chr20_43104144_43104478 | 0.11 |
TTPAL |
tocopherol (alpha) transfer protein-like |
215 |
0.89 |
| chrX_55282426_55282577 | 0.11 |
PAGE3 |
P antigen family, member 3 (prostate associated) |
8662 |
0.21 |
| chr1_33472964_33473115 | 0.11 |
RP1-117O3.2 |
|
20363 |
0.13 |
| chr8_126398781_126398932 | 0.11 |
RP11-550A5.2 |
|
35198 |
0.17 |
| chr10_45956119_45956270 | 0.11 |
RP11-67C2.2 |
|
7625 |
0.26 |
| chr3_46508059_46508210 | 0.11 |
LTF |
lactotransferrin |
1481 |
0.36 |
| chr5_76361780_76361931 | 0.11 |
CTC-564N23.2 |
|
7851 |
0.14 |
| chr7_128580814_128580965 | 0.11 |
IRF5 |
interferon regulatory factor 5 |
163 |
0.94 |
| chr2_136871413_136871564 | 0.11 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
2325 |
0.41 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0052308 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0042520 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |