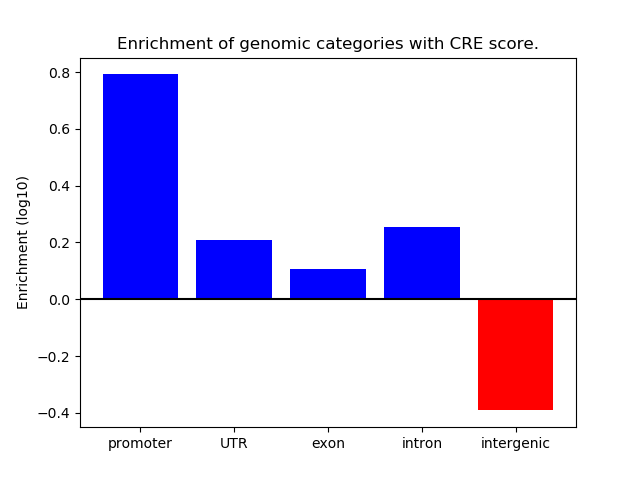
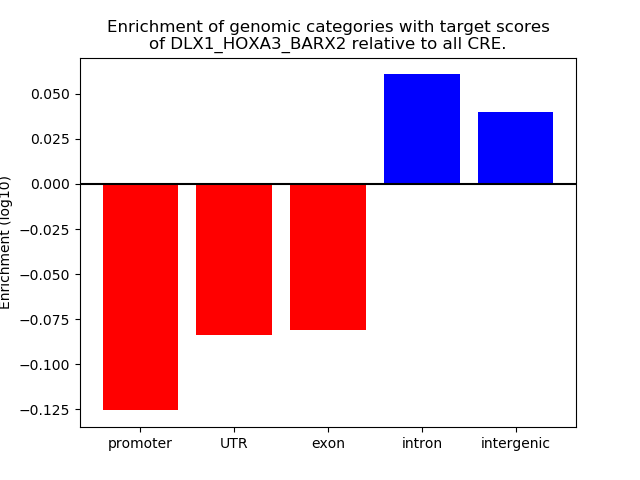
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
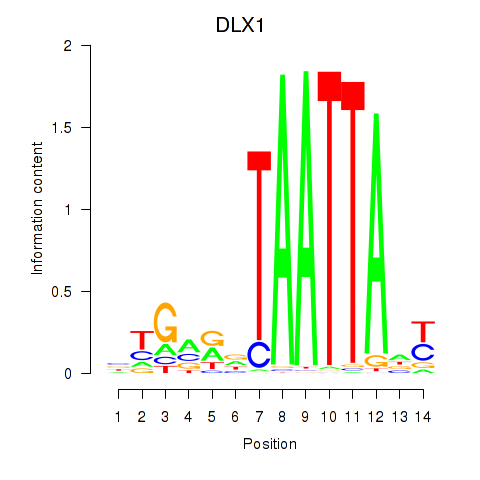
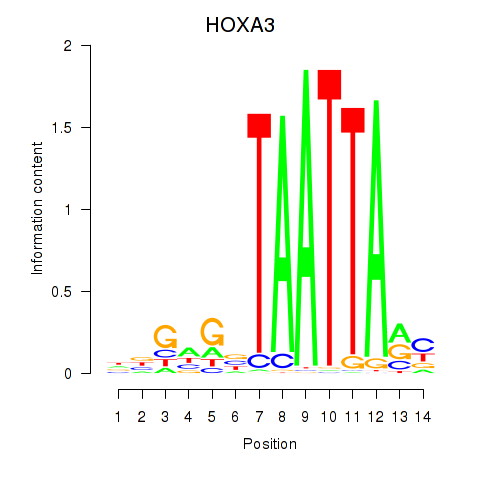
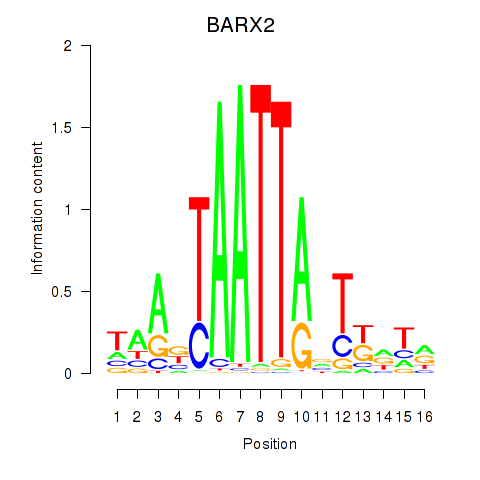
Results for DLX1_HOXA3_BARX2
Z-value: 1.02
Transcription factors associated with DLX1_HOXA3_BARX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX1
|
ENSG00000144355.10 | distal-less homeobox 1 |
|
HOXA3
|
ENSG00000105997.18 | homeobox A3 |
|
BARX2
|
ENSG00000043039.5 | BARX homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_129245088_129245270 | BARX2 | 656 | 0.816313 | 0.56 | 1.2e-01 | Click! |
| chr11_129243062_129243656 | BARX2 | 2476 | 0.412366 | 0.45 | 2.3e-01 | Click! |
| chr11_129242275_129242426 | BARX2 | 3485 | 0.356232 | -0.41 | 2.7e-01 | Click! |
| chr11_129244074_129244550 | BARX2 | 1523 | 0.542584 | 0.32 | 4.0e-01 | Click! |
| chr11_129242593_129242759 | BARX2 | 3159 | 0.369655 | 0.32 | 4.0e-01 | Click! |
| chr2_172951434_172951585 | DLX1 | 1245 | 0.496892 | 0.77 | 1.5e-02 | Click! |
| chr2_172948607_172948758 | DLX1 | 786 | 0.674454 | 0.77 | 1.6e-02 | Click! |
| chr2_172958009_172958160 | DLX1 | 7820 | 0.207962 | 0.70 | 3.5e-02 | Click! |
| chr2_172947668_172947819 | DLX1 | 1725 | 0.393210 | 0.63 | 6.9e-02 | Click! |
| chr2_172917249_172917400 | DLX1 | 32144 | 0.164637 | 0.52 | 1.5e-01 | Click! |
| chr7_27150712_27150972 | HOXA3 | 2622 | 0.104312 | 0.75 | 2.0e-02 | Click! |
| chr7_27152154_27152305 | HOXA3 | 1235 | 0.194179 | 0.64 | 6.4e-02 | Click! |
| chr7_27150237_27150691 | HOXA3 | 3000 | 0.096485 | 0.62 | 7.5e-02 | Click! |
| chr7_27176258_27176409 | HOXA3 | 3492 | 0.077544 | 0.61 | 7.9e-02 | Click! |
| chr7_27151802_27151953 | HOXA3 | 1587 | 0.153829 | 0.57 | 1.1e-01 | Click! |
Activity of the DLX1_HOXA3_BARX2 motif across conditions
Conditions sorted by the z-value of the DLX1_HOXA3_BARX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_87036535_87037613 | 1.60 |
VGLL3 |
vestigial like 3 (Drosophila) |
2778 |
0.42 |
| chr7_129933276_129933885 | 1.58 |
CPA4 |
carboxypeptidase A4 |
461 |
0.76 |
| chr7_93550787_93551002 | 1.27 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
117 |
0.96 |
| chr21_17909068_17909746 | 1.11 |
ENSG00000207638 |
. |
2002 |
0.33 |
| chr7_95107815_95108198 | 1.06 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
7207 |
0.2 |
| chr8_116675050_116675578 | 1.03 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr3_105657205_105657471 | 1.02 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
68942 |
0.15 |
| chr13_24146066_24146217 | 0.98 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1338 |
0.59 |
| chr4_77506894_77507309 | 0.95 |
ENSG00000265314 |
. |
10397 |
0.17 |
| chr9_18474095_18474958 | 0.91 |
ADAMTSL1 |
ADAMTS-like 1 |
295 |
0.95 |
| chr3_33318777_33319866 | 0.89 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
353 |
0.91 |
| chr7_17720120_17720505 | 0.89 |
SNX13 |
sorting nexin 13 |
259779 |
0.02 |
| chr11_131780111_131780399 | 0.87 |
NTM |
neurotrimin |
642 |
0.77 |
| chr8_93114852_93115389 | 0.86 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
334 |
0.94 |
| chr6_116833589_116833880 | 0.84 |
TRAPPC3L |
trafficking protein particle complex 3-like |
228 |
0.83 |
| chr13_73081776_73081993 | 0.83 |
ENSG00000251715 |
. |
47145 |
0.15 |
| chr18_53089837_53090372 | 0.83 |
TCF4 |
transcription factor 4 |
361 |
0.89 |
| chr6_126135984_126136135 | 0.79 |
NCOA7 |
nuclear receptor coactivator 7 |
305 |
0.89 |
| chr20_50384318_50384901 | 0.79 |
ATP9A |
ATPase, class II, type 9A |
258 |
0.94 |
| chr7_93552303_93552503 | 0.78 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
1392 |
0.38 |
| chr12_66286178_66286489 | 0.76 |
RP11-366L20.2 |
Uncharacterized protein |
10386 |
0.18 |
| chr1_196577911_196578299 | 0.76 |
KCNT2 |
potassium channel, subfamily T, member 2 |
250 |
0.94 |
| chr18_53090404_53090609 | 0.76 |
TCF4 |
transcription factor 4 |
763 |
0.7 |
| chr4_57975072_57975270 | 0.75 |
IGFBP7-AS1 |
IGFBP7 antisense RNA 1 |
757 |
0.52 |
| chr6_85823864_85824548 | 0.75 |
NT5E |
5'-nucleotidase, ecto (CD73) |
335603 |
0.01 |
| chr17_19998529_19998923 | 0.74 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
279 |
0.92 |
| chr11_12849556_12849989 | 0.73 |
RP11-47J17.3 |
|
4558 |
0.23 |
| chr12_66220556_66220804 | 0.73 |
HMGA2 |
high mobility group AT-hook 2 |
1777 |
0.39 |
| chr12_56917781_56918152 | 0.73 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
2183 |
0.23 |
| chr12_78336292_78336638 | 0.72 |
NAV3 |
neuron navigator 3 |
23591 |
0.28 |
| chr12_66221036_66221187 | 0.69 |
HMGA2 |
high mobility group AT-hook 2 |
2208 |
0.34 |
| chr15_37179413_37179956 | 0.69 |
ENSG00000212511 |
. |
34843 |
0.22 |
| chr4_134070427_134071051 | 0.68 |
PCDH10 |
protocadherin 10 |
269 |
0.96 |
| chr1_68697735_68698156 | 0.68 |
WLS |
wntless Wnt ligand secretion mediator |
18 |
0.98 |
| chr3_112356169_112356393 | 0.68 |
CCDC80 |
coiled-coil domain containing 80 |
663 |
0.77 |
| chr6_19691587_19692480 | 0.68 |
ENSG00000200957 |
. |
49273 |
0.18 |
| chr8_38855409_38855789 | 0.67 |
ADAM9 |
ADAM metallopeptidase domain 9 |
1094 |
0.34 |
| chr1_84107245_84107582 | 0.67 |
ENSG00000223231 |
. |
152147 |
0.04 |
| chr10_60273912_60274168 | 0.66 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
1140 |
0.65 |
| chr13_38172411_38172686 | 0.66 |
POSTN |
periostin, osteoblast specific factor |
315 |
0.95 |
| chr3_79066759_79066910 | 0.65 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
860 |
0.75 |
| chr2_207998546_207999225 | 0.65 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
24 |
0.98 |
| chr15_96878465_96878729 | 0.65 |
ENSG00000222651 |
. |
2107 |
0.24 |
| chr8_37350646_37351319 | 0.64 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr1_87799796_87800719 | 0.64 |
LMO4 |
LIM domain only 4 |
2906 |
0.4 |
| chr2_151340168_151340387 | 0.63 |
RND3 |
Rho family GTPase 3 |
1619 |
0.57 |
| chr16_54968513_54968925 | 0.63 |
IRX5 |
iroquois homeobox 5 |
2735 |
0.43 |
| chr20_45946936_45948261 | 0.63 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
352 |
0.82 |
| chr4_41215270_41215769 | 0.61 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
956 |
0.59 |
| chr5_66311217_66311678 | 0.61 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
10964 |
0.24 |
| chr15_99794226_99794420 | 0.61 |
LRRC28 |
leucine rich repeat containing 28 |
2307 |
0.22 |
| chr5_88080560_88081020 | 0.61 |
MEF2C |
myocyte enhancer factor 2C |
38815 |
0.18 |
| chr10_6763782_6764322 | 0.61 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr20_10502111_10502382 | 0.61 |
SLX4IP |
SLX4 interacting protein |
86295 |
0.09 |
| chr5_174178214_174178699 | 0.61 |
ENSG00000266890 |
. |
281 |
0.94 |
| chr11_111849006_111849202 | 0.61 |
DIXDC1 |
DIX domain containing 1 |
1071 |
0.39 |
| chr10_86185479_86185663 | 0.60 |
CCSER2 |
coiled-coil serine-rich protein 2 |
876 |
0.74 |
| chr4_157997366_157997978 | 0.60 |
GLRB |
glycine receptor, beta |
117 |
0.97 |
| chr3_114477857_114478359 | 0.60 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
10 |
0.99 |
| chr11_106239153_106239583 | 0.60 |
RP11-680E19.1 |
|
104326 |
0.08 |
| chr17_66700653_66700804 | 0.60 |
ENSG00000263690 |
. |
61972 |
0.13 |
| chr18_20612011_20612377 | 0.59 |
ENSG00000223023 |
. |
7635 |
0.17 |
| chr2_38295840_38295991 | 0.59 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
1731 |
0.34 |
| chr3_110791913_110792064 | 0.59 |
PVRL3 |
poliovirus receptor-related 3 |
973 |
0.53 |
| chr5_149867786_149868254 | 0.59 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
2163 |
0.3 |
| chr15_42749459_42749764 | 0.58 |
ZNF106 |
zinc finger protein 106 |
100 |
0.96 |
| chr8_57086542_57086822 | 0.58 |
PLAG1 |
pleiomorphic adenoma gene 1 |
37156 |
0.12 |
| chr2_58266503_58266768 | 0.57 |
VRK2 |
vaccinia related kinase 2 |
7094 |
0.27 |
| chr4_157891142_157892336 | 0.57 |
PDGFC |
platelet derived growth factor C |
316 |
0.91 |
| chr2_200446222_200446758 | 0.57 |
SATB2 |
SATB homeobox 2 |
110501 |
0.07 |
| chr2_66668625_66669005 | 0.57 |
AC092669.1 |
|
153 |
0.89 |
| chr4_82413627_82413778 | 0.56 |
RASGEF1B |
RasGEF domain family, member 1B |
20633 |
0.28 |
| chr4_15471989_15472140 | 0.56 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
427 |
0.85 |
| chr10_101736542_101736753 | 0.56 |
DNMBP |
dynamin binding protein |
33029 |
0.15 |
| chr20_43965593_43965922 | 0.56 |
SDC4 |
syndecan 4 |
11307 |
0.11 |
| chr6_27060216_27060434 | 0.56 |
ENSG00000222800 |
. |
17434 |
0.13 |
| chr14_29859562_29859789 | 0.56 |
ENSG00000257522 |
. |
11813 |
0.29 |
| chr5_82767889_82768074 | 0.55 |
VCAN |
versican |
237 |
0.96 |
| chr9_104248314_104249564 | 0.55 |
TMEM246 |
transmembrane protein 246 |
460 |
0.79 |
| chr15_88020923_88021173 | 0.55 |
ENSG00000207150 |
. |
3528 |
0.4 |
| chr21_36253539_36254227 | 0.55 |
RUNX1 |
runt-related transcription factor 1 |
5597 |
0.33 |
| chr4_108746841_108747185 | 0.55 |
SGMS2 |
sphingomyelin synthase 2 |
649 |
0.76 |
| chr9_89952337_89952906 | 0.55 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr22_36233928_36234199 | 0.55 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
2202 |
0.43 |
| chr15_80365541_80365692 | 0.55 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
684 |
0.75 |
| chr3_114169976_114170457 | 0.54 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
3314 |
0.33 |
| chr15_71054590_71054766 | 0.54 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
1091 |
0.55 |
| chr1_39881370_39881756 | 0.54 |
MACF1 |
microtubule-actin crosslinking factor 1 |
1491 |
0.39 |
| chr3_61546098_61546562 | 0.54 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
1255 |
0.64 |
| chr6_163817634_163818149 | 0.54 |
QKI |
QKI, KH domain containing, RNA binding |
17784 |
0.29 |
| chr1_162603392_162603656 | 0.54 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
1264 |
0.48 |
| chr5_58592476_58593460 | 0.54 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
21023 |
0.29 |
| chr1_170640402_170640573 | 0.54 |
PRRX1 |
paired related homeobox 1 |
7409 |
0.3 |
| chr8_131206424_131206650 | 0.54 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
13429 |
0.23 |
| chr1_68214173_68214559 | 0.54 |
ENSG00000238778 |
. |
23970 |
0.19 |
| chr12_89845524_89845923 | 0.54 |
POC1B |
POC1 centriolar protein B |
45311 |
0.13 |
| chr6_86167890_86168219 | 0.53 |
NT5E |
5'-nucleotidase, ecto (CD73) |
8227 |
0.28 |
| chr11_121966638_121966789 | 0.53 |
ENSG00000207971 |
. |
3839 |
0.19 |
| chr4_71479096_71479386 | 0.53 |
ENAM |
enamelin |
15220 |
0.15 |
| chr10_69831641_69832005 | 0.53 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
2071 |
0.33 |
| chr15_99864758_99865035 | 0.53 |
AC022819.2 |
Uncharacterized protein |
5068 |
0.23 |
| chr17_35851874_35852313 | 0.53 |
DUSP14 |
dual specificity phosphatase 14 |
523 |
0.79 |
| chr14_101867418_101867569 | 0.53 |
ENSG00000258498 |
. |
159266 |
0.02 |
| chr13_33858125_33858279 | 0.53 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
1690 |
0.4 |
| chr4_6887879_6888223 | 0.52 |
TBC1D14 |
TBC1 domain family, member 14 |
22918 |
0.18 |
| chr7_27202641_27202952 | 0.52 |
HOXA9 |
homeobox A9 |
2349 |
0.09 |
| chr4_124339787_124340099 | 0.52 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
18820 |
0.3 |
| chr16_53244996_53245358 | 0.52 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
2814 |
0.29 |
| chr10_33620428_33620906 | 0.52 |
NRP1 |
neuropilin 1 |
2643 |
0.37 |
| chr8_62625183_62625719 | 0.52 |
ASPH |
aspartate beta-hydroxylase |
1633 |
0.37 |
| chr8_77599205_77599356 | 0.51 |
ZFHX4 |
zinc finger homeobox 4 |
3264 |
0.28 |
| chr7_14028526_14029745 | 0.51 |
ETV1 |
ets variant 1 |
156 |
0.97 |
| chr13_76588981_76589230 | 0.51 |
ENSG00000243274 |
. |
119962 |
0.06 |
| chr1_89447034_89447185 | 0.51 |
CCBL2 |
cysteine conjugate-beta lyase 2 |
11178 |
0.15 |
| chr2_145210039_145210510 | 0.51 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
22137 |
0.25 |
| chr3_79411197_79411348 | 0.51 |
ENSG00000265193 |
. |
145765 |
0.05 |
| chr2_110656312_110656910 | 0.51 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
343 |
0.93 |
| chr10_115032925_115033406 | 0.51 |
ENSG00000238380 |
. |
80019 |
0.11 |
| chr3_120215962_120216316 | 0.51 |
FSTL1 |
follistatin-like 1 |
46039 |
0.16 |
| chr6_26577102_26577344 | 0.51 |
ABT1 |
activator of basal transcription 1 |
19957 |
0.15 |
| chr1_153585227_153585451 | 0.51 |
S100A16 |
S100 calcium binding protein A16 |
220 |
0.83 |
| chr8_108506884_108507225 | 0.51 |
ANGPT1 |
angiopoietin 1 |
169 |
0.98 |
| chr9_79483882_79484033 | 0.51 |
PRUNE2 |
prune homolog 2 (Drosophila) |
37044 |
0.21 |
| chrX_114829080_114829277 | 0.50 |
PLS3 |
plastin 3 |
1313 |
0.48 |
| chr3_44063258_44064101 | 0.50 |
ENSG00000252980 |
. |
48900 |
0.17 |
| chr9_22008020_22008450 | 0.50 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
717 |
0.61 |
| chr3_174159101_174159924 | 0.50 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
735 |
0.81 |
| chr4_54063057_54063462 | 0.49 |
ENSG00000207385 |
. |
68475 |
0.13 |
| chr5_158524974_158525234 | 0.49 |
EBF1 |
early B-cell factor 1 |
1597 |
0.48 |
| chr16_73089798_73090050 | 0.49 |
ZFHX3 |
zinc finger homeobox 3 |
3673 |
0.28 |
| chr14_75039149_75039311 | 0.49 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
39545 |
0.13 |
| chr5_107229567_107229740 | 0.49 |
ENSG00000251732 |
. |
83323 |
0.11 |
| chr1_151918063_151918251 | 0.48 |
THEM4 |
thioesterase superfamily member 4 |
35873 |
0.1 |
| chr2_109271526_109272143 | 0.48 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
325 |
0.91 |
| chr2_128492018_128492224 | 0.48 |
SFT2D3 |
SFT2 domain containing 3 |
33524 |
0.13 |
| chr2_198540490_198540682 | 0.48 |
RFTN2 |
raftlin family member 2 |
133 |
0.96 |
| chr17_42214986_42215221 | 0.48 |
ENSG00000212446 |
. |
1746 |
0.18 |
| chr3_116583114_116583333 | 0.48 |
ENSG00000265433 |
. |
14009 |
0.25 |
| chrX_16825753_16826181 | 0.48 |
TXLNG |
taxilin gamma |
21412 |
0.16 |
| chr20_56749874_56750278 | 0.48 |
C20orf85 |
chromosome 20 open reading frame 85 |
24116 |
0.19 |
| chr18_56244152_56244311 | 0.48 |
RP11-126O1.2 |
|
19929 |
0.14 |
| chr7_151297260_151297484 | 0.48 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
32067 |
0.18 |
| chr6_28442401_28443147 | 0.48 |
ZSCAN23 |
zinc finger and SCAN domain containing 23 |
31530 |
0.13 |
| chr8_77589944_77590318 | 0.48 |
ZFHX4 |
zinc finger homeobox 4 |
3323 |
0.29 |
| chr8_48648492_48649280 | 0.48 |
CEBPD |
CCAAT/enhancer binding protein (C/EBP), delta |
2762 |
0.26 |
| chr7_151328064_151328292 | 0.47 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
1261 |
0.55 |
| chr6_148829582_148830289 | 0.47 |
ENSG00000223322 |
. |
15441 |
0.29 |
| chr5_111090218_111090459 | 0.47 |
NREP |
neuronal regeneration related protein |
1610 |
0.42 |
| chr6_165341513_165341675 | 0.47 |
C6orf118 |
chromosome 6 open reading frame 118 |
380748 |
0.01 |
| chr7_28726429_28726958 | 0.47 |
CREB5 |
cAMP responsive element binding protein 5 |
1095 |
0.68 |
| chr14_53509566_53509717 | 0.47 |
RP11-368P15.3 |
|
6168 |
0.27 |
| chr6_27859319_27859470 | 0.47 |
HIST1H3J |
histone cluster 1, H3j |
824 |
0.31 |
| chr12_120246427_120246712 | 0.47 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
5382 |
0.27 |
| chr6_5471426_5471612 | 0.47 |
RP1-232P20.1 |
|
13211 |
0.27 |
| chr3_59101471_59101696 | 0.47 |
C3orf67 |
chromosome 3 open reading frame 67 |
65773 |
0.15 |
| chr10_73725086_73725706 | 0.47 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
1273 |
0.55 |
| chr6_1613704_1614304 | 0.46 |
FOXC1 |
forkhead box C1 |
3323 |
0.37 |
| chr1_79506717_79506893 | 0.46 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
34402 |
0.23 |
| chr15_80840560_80840711 | 0.46 |
RP11-379K22.2 |
|
14650 |
0.18 |
| chr11_114167823_114168031 | 0.46 |
NNMT |
nicotinamide N-methyltransferase |
158 |
0.96 |
| chr5_3597777_3597928 | 0.46 |
IRX1 |
iroquois homeobox 1 |
1684 |
0.41 |
| chr19_1361544_1361801 | 0.46 |
MUM1 |
melanoma associated antigen (mutated) 1 |
5309 |
0.1 |
| chr12_47470653_47470804 | 0.46 |
PCED1B |
PC-esterase domain containing 1B |
2658 |
0.29 |
| chr13_37006714_37007207 | 0.46 |
CCNA1 |
cyclin A1 |
465 |
0.85 |
| chr5_158488269_158488591 | 0.46 |
EBF1 |
early B-cell factor 1 |
38271 |
0.17 |
| chr12_22059820_22060034 | 0.46 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
3985 |
0.3 |
| chr9_91146971_91147229 | 0.46 |
NXNL2 |
nucleoredoxin-like 2 |
2916 |
0.39 |
| chr7_98970111_98970529 | 0.45 |
ARPC1B |
actin related protein 2/3 complex, subunit 1B, 41kDa |
1552 |
0.28 |
| chr22_36232810_36232961 | 0.45 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3380 |
0.35 |
| chr12_94580963_94581249 | 0.45 |
RP11-74K11.2 |
|
1286 |
0.5 |
| chr13_76217437_76217800 | 0.45 |
LMO7 |
LIM domain 7 |
7159 |
0.18 |
| chr8_107738861_107739281 | 0.45 |
OXR1 |
oxidation resistance 1 |
658 |
0.71 |
| chr4_109532705_109532935 | 0.45 |
RPL34 |
ribosomal protein L34 |
8902 |
0.2 |
| chr5_14153831_14154379 | 0.45 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10276 |
0.32 |
| chr7_134396917_134397381 | 0.45 |
CALD1 |
caldesmon 1 |
31854 |
0.2 |
| chr5_16916952_16917142 | 0.45 |
MYO10 |
myosin X |
411 |
0.87 |
| chr6_56705417_56705587 | 0.45 |
DST |
dystonin |
2441 |
0.31 |
| chr4_48700842_48701028 | 0.45 |
FRYL |
FRY-like |
17747 |
0.24 |
| chr16_89990965_89991193 | 0.45 |
TUBB3 |
Tubulin beta-3 chain |
1305 |
0.26 |
| chr18_60117928_60118203 | 0.45 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
72175 |
0.1 |
| chr1_38910382_38910533 | 0.44 |
ENSG00000200796 |
. |
48126 |
0.18 |
| chr2_159651901_159652191 | 0.44 |
DAPL1 |
death associated protein-like 1 |
217 |
0.95 |
| chr20_46412966_46413363 | 0.44 |
SULF2 |
sulfatase 2 |
1069 |
0.59 |
| chr4_95855683_95855842 | 0.44 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
61185 |
0.16 |
| chr12_94385101_94385284 | 0.44 |
ENSG00000223126 |
. |
17631 |
0.22 |
| chr8_72273557_72273741 | 0.44 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
446 |
0.9 |
| chr5_173000019_173000195 | 0.44 |
CTB-33O18.3 |
|
6539 |
0.26 |
| chr2_46933523_46933944 | 0.44 |
SOCS5 |
suppressor of cytokine signaling 5 |
7407 |
0.22 |
| chr11_122311553_122311819 | 0.44 |
ENSG00000252776 |
. |
8232 |
0.23 |
| chr1_109939510_109939955 | 0.44 |
SORT1 |
sortilin 1 |
841 |
0.56 |
| chr9_112955081_112955242 | 0.44 |
C9orf152 |
chromosome 9 open reading frame 152 |
15308 |
0.23 |
| chr6_116834038_116834189 | 0.43 |
TRAPPC3L |
trafficking protein particle complex 3-like |
607 |
0.55 |
| chr9_103192985_103193136 | 0.43 |
MSANTD3 |
Myb/SANT-like DNA-binding domain containing 3 |
3341 |
0.21 |
| chr1_169883940_169884437 | 0.43 |
SCYL3 |
SCY1-like 3 (S. cerevisiae) |
20780 |
0.18 |
| chr2_64877806_64878096 | 0.43 |
SERTAD2 |
SERTA domain containing 2 |
3096 |
0.31 |
| chr2_188445720_188445877 | 0.43 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
15311 |
0.23 |
| chr18_42313149_42313495 | 0.43 |
SETBP1 |
SET binding protein 1 |
36193 |
0.24 |
| chr1_95330057_95330663 | 0.43 |
SLC44A3 |
solute carrier family 44, member 3 |
2527 |
0.28 |
| chr11_31830603_31830754 | 0.43 |
PAX6 |
paired box 6 |
1175 |
0.49 |
| chr12_118628413_118628612 | 0.43 |
TAOK3 |
TAO kinase 3 |
156 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 | 1.3 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.4 | 1.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.3 | 1.2 | GO:0060013 | righting reflex(GO:0060013) |
| 0.3 | 0.9 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.6 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.2 | 0.6 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.2 | 0.9 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.2 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 0.5 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.2 | 0.5 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.2 | 0.5 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.2 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 1.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.4 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.4 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.1 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 0.4 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.5 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.1 | 0.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.8 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.1 | 0.5 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.2 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.1 | 0.3 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 0.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.3 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.1 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.4 | GO:0010659 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.1 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 0.1 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.1 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.9 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.1 | GO:0061140 | lung secretory cell differentiation(GO:0061140) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.5 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.1 | 1.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.2 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.1 | 0.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.2 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.3 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.1 | 0.3 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.1 | 1.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 0.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 1.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.1 | 0.2 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.1 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.1 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 1.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0009415 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.1 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.2 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.6 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 0.3 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.1 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.1 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 1.3 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.1 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.3 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.1 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.9 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.7 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.2 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 1.1 | GO:0060606 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:1903504 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.7 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.4 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0032616 | interleukin-13 production(GO:0032616) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 1.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0042634 | regulation of hair cycle(GO:0042634) regulation of hair follicle development(GO:0051797) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0045628 | regulation of T-helper 2 cell differentiation(GO:0045628) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.3 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 0.7 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:1904063 | negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.3 | GO:0047496 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0031935 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0034444 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.2 | GO:0046036 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0097035 | phospholipid scrambling(GO:0017121) regulation of membrane lipid distribution(GO:0097035) |
| 0.0 | 0.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 3.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0051307 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.5 | GO:0046068 | cGMP metabolic process(GO:0046068) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.5 | GO:0007215 | glutamate receptor signaling pathway(GO:0007215) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.3 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.3 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.0 | GO:0045188 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0032106 | positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 0.2 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.3 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.0 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.3 | GO:0030318 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 0.1 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.0 | GO:0051955 | regulation of amino acid transport(GO:0051955) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:1904181 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.0 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.0 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.0 | 0.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.0 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.5 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.0 | GO:0044319 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0042461 | photoreceptor cell development(GO:0042461) eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.0 | GO:1902913 | positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0035510 | DNA dealkylation(GO:0035510) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.0 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.6 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.2 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.4 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.8 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0044217 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.1 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.0 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.0 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.3 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.7 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.2 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.4 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.6 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.1 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.6 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.4 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.3 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.1 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.9 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.1 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.2 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 2.8 | GO:0030528 | obsolete transcription regulator activity(GO:0030528) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 1.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.9 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.8 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |