Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
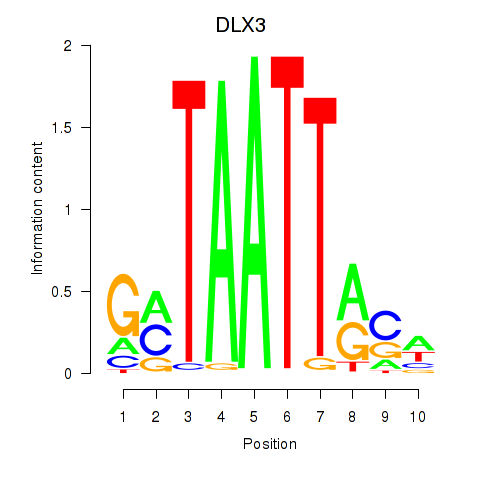
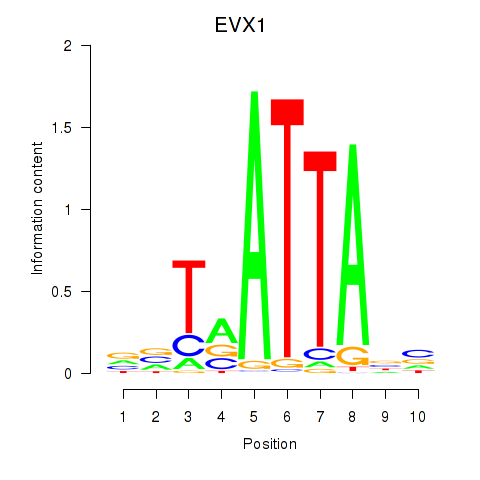
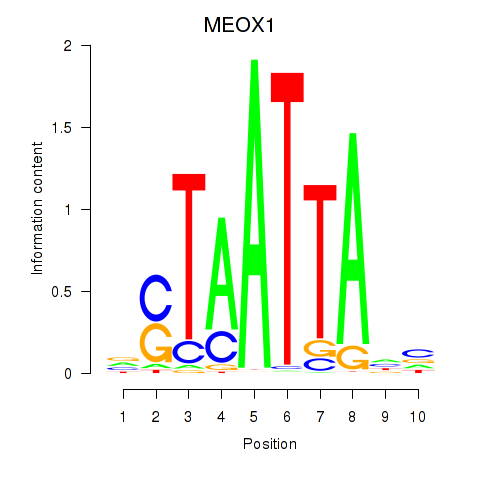
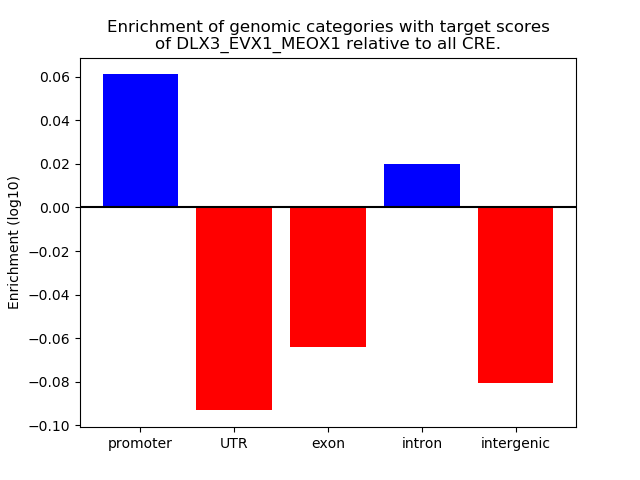
Results for DLX3_EVX1_MEOX1
Z-value: 0.47
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX3
|
ENSG00000064195.7 | distal-less homeobox 3 |
|
EVX1
|
ENSG00000106038.8 | even-skipped homeobox 1 |
|
MEOX1
|
ENSG00000005102.8 | mesenchyme homeobox 1 |
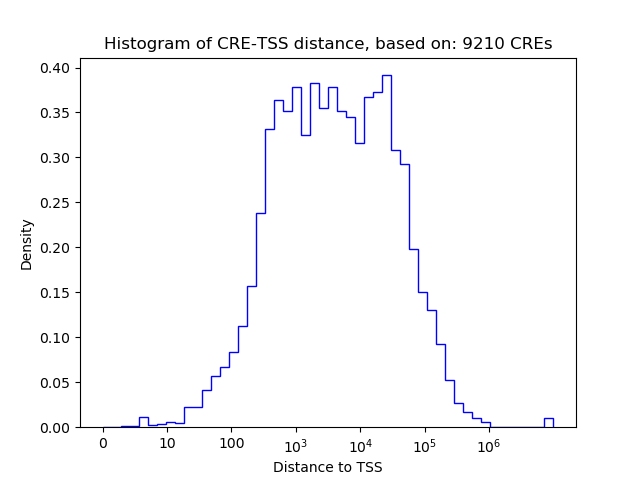
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_48074174_48074657 | DLX3 | 1827 | 0.283554 | -0.71 | 3.3e-02 | Click! |
| chr17_48071900_48072168 | DLX3 | 554 | 0.711802 | -0.68 | 4.2e-02 | Click! |
| chr17_48065551_48065702 | DLX3 | 5441 | 0.160340 | -0.63 | 6.8e-02 | Click! |
| chr17_48075878_48076029 | DLX3 | 3365 | 0.191350 | -0.62 | 7.7e-02 | Click! |
| chr17_48072265_48072578 | DLX3 | 167 | 0.938338 | -0.60 | 8.9e-02 | Click! |
| chr7_27275476_27275737 | EVX1 | 6558 | 0.102992 | -0.78 | 1.2e-02 | Click! |
| chr7_27282525_27283078 | EVX1 | 186 | 0.525290 | -0.74 | 2.2e-02 | Click! |
| chr7_27274920_27275071 | EVX1 | 7169 | 0.100761 | -0.69 | 3.9e-02 | Click! |
| chr7_27275140_27275379 | EVX1 | 6905 | 0.101684 | -0.63 | 6.9e-02 | Click! |
| chr7_27282158_27282489 | EVX1 | 4 | 0.772250 | -0.57 | 1.1e-01 | Click! |
| chr17_41712240_41712391 | MEOX1 | 26616 | 0.143953 | -0.64 | 6.1e-02 | Click! |
| chr17_41736574_41736725 | MEOX1 | 2282 | 0.286119 | 0.59 | 9.2e-02 | Click! |
| chr17_41738065_41738244 | MEOX1 | 777 | 0.637485 | 0.52 | 1.5e-01 | Click! |
| chr17_41737286_41737476 | MEOX1 | 1550 | 0.380643 | 0.50 | 1.7e-01 | Click! |
| chr17_41737840_41737991 | MEOX1 | 1016 | 0.530961 | 0.44 | 2.4e-01 | Click! |
Activity of the DLX3_EVX1_MEOX1 motif across conditions
Conditions sorted by the z-value of the DLX3_EVX1_MEOX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
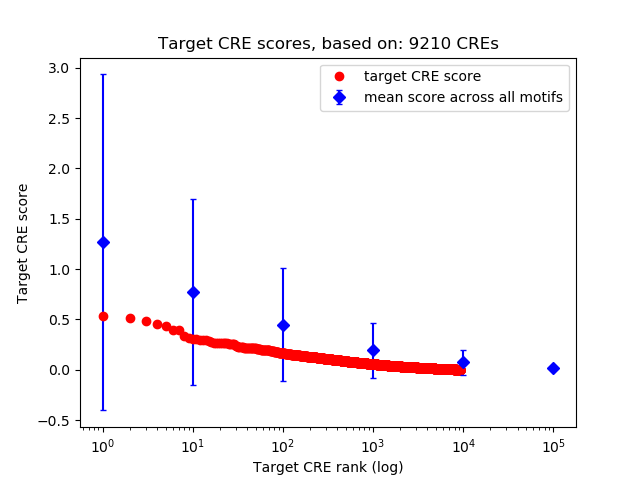
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_78400540_78401325 | 0.53 |
GPR174 |
G protein-coupled receptor 174 |
25537 |
0.27 |
| chr6_154568354_154568864 | 0.52 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
| chr21_15917916_15918619 | 0.48 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr13_99957828_99957979 | 0.45 |
GPR183 |
G protein-coupled receptor 183 |
1756 |
0.38 |
| chr15_75080868_75081404 | 0.44 |
ENSG00000264386 |
. |
38 |
0.76 |
| chr2_158299843_158300108 | 0.39 |
CYTIP |
cytohesin 1 interacting protein |
679 |
0.68 |
| chr6_24934739_24934890 | 0.39 |
FAM65B |
family with sequence similarity 65, member B |
1374 |
0.5 |
| chr11_122597673_122598179 | 0.34 |
ENSG00000239079 |
. |
907 |
0.67 |
| chr12_866633_866784 | 0.31 |
WNK1 |
WNK lysine deficient protein kinase 1 |
3976 |
0.25 |
| chr20_43599792_43600332 | 0.31 |
STK4 |
serine/threonine kinase 4 |
4895 |
0.16 |
| chr2_143887292_143887566 | 0.30 |
ARHGAP15 |
Rho GTPase activating protein 15 |
546 |
0.84 |
| chr5_149791368_149791565 | 0.29 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
829 |
0.6 |
| chr1_234544831_234544982 | 0.29 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
35459 |
0.12 |
| chr11_75063727_75063930 | 0.29 |
ARRB1 |
arrestin, beta 1 |
955 |
0.48 |
| chr14_22993528_22993679 | 0.29 |
TRAJ15 |
T cell receptor alpha joining 15 |
4977 |
0.12 |
| chr13_40532306_40532606 | 0.27 |
ENSG00000212553 |
. |
101092 |
0.08 |
| chr10_6626003_6626633 | 0.27 |
PRKCQ |
protein kinase C, theta |
4055 |
0.36 |
| chrX_78200973_78201449 | 0.27 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
293 |
0.95 |
| chr15_55547653_55548107 | 0.27 |
RAB27A |
RAB27A, member RAS oncogene family |
6647 |
0.22 |
| chr7_150329820_150329971 | 0.27 |
GIMAP6 |
GTPase, IMAP family member 6 |
422 |
0.84 |
| chr2_136876136_136876323 | 0.26 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
494 |
0.87 |
| chr9_95730621_95730837 | 0.26 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
4486 |
0.25 |
| chr17_10601145_10601947 | 0.26 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
635 |
0.41 |
| chr12_54891959_54892179 | 0.26 |
NCKAP1L |
NCK-associated protein 1-like |
499 |
0.73 |
| chr4_154411300_154411567 | 0.26 |
KIAA0922 |
KIAA0922 |
23932 |
0.22 |
| chrX_147462854_147463013 | 0.26 |
AC002368.4 |
|
119202 |
0.06 |
| chr10_73847731_73848399 | 0.26 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
21 |
0.98 |
| chr13_21050204_21050392 | 0.25 |
ENSG00000263978 |
. |
42313 |
0.15 |
| chr8_82022711_82022967 | 0.25 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
1464 |
0.56 |
| chr13_77319930_77320170 | 0.24 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
140475 |
0.05 |
| chr2_182174133_182174359 | 0.24 |
ENSG00000266705 |
. |
3867 |
0.36 |
| chr14_22955963_22956254 | 0.23 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
63 |
0.95 |
| chr19_49139505_49139656 | 0.23 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
1040 |
0.28 |
| chr3_152878700_152878913 | 0.23 |
RP11-529G21.2 |
|
758 |
0.56 |
| chr6_32158116_32158500 | 0.23 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
345 |
0.7 |
| chr2_225809390_225809894 | 0.22 |
DOCK10 |
dedicator of cytokinesis 10 |
2140 |
0.45 |
| chr9_95790679_95791025 | 0.22 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
13510 |
0.18 |
| chr12_27150397_27150548 | 0.22 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
2271 |
0.24 |
| chr12_131796680_131796831 | 0.22 |
ENSG00000212251 |
. |
12459 |
0.23 |
| chr10_129861850_129862130 | 0.22 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
16156 |
0.26 |
| chr2_88124610_88124761 | 0.22 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
601 |
0.82 |
| chr17_75456326_75456913 | 0.22 |
SEPT9 |
septin 9 |
4171 |
0.18 |
| chr5_110560941_110561277 | 0.22 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1325 |
0.51 |
| chrX_40430227_40430433 | 0.22 |
ATP6AP2 |
ATPase, H+ transporting, lysosomal accessory protein 2 |
9816 |
0.21 |
| chr10_28622520_28622887 | 0.22 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
712 |
0.72 |
| chr3_16883621_16883873 | 0.22 |
PLCL2 |
phospholipase C-like 2 |
42705 |
0.19 |
| chr6_70525806_70525957 | 0.22 |
LMBRD1 |
LMBR1 domain containing 1 |
18878 |
0.25 |
| chr2_161995560_161995711 | 0.21 |
TANK |
TRAF family member-associated NFKB activator |
2169 |
0.37 |
| chr8_56776921_56777072 | 0.21 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
15376 |
0.15 |
| chr5_126094279_126094440 | 0.21 |
ENSG00000252185 |
. |
3250 |
0.29 |
| chr6_15308796_15308947 | 0.21 |
ENSG00000201367 |
. |
6280 |
0.22 |
| chr3_151921407_151921835 | 0.21 |
MBNL1 |
muscleblind-like splicing regulator 1 |
64208 |
0.12 |
| chr2_68993241_68993423 | 0.21 |
ARHGAP25 |
Rho GTPase activating protein 25 |
8601 |
0.25 |
| chr5_14560333_14560610 | 0.21 |
FAM105A |
family with sequence similarity 105, member A |
21413 |
0.23 |
| chr7_37011253_37011404 | 0.21 |
ELMO1 |
engulfment and cell motility 1 |
13337 |
0.2 |
| chr4_109056842_109056993 | 0.20 |
LEF1 |
lymphoid enhancer-binding factor 1 |
30540 |
0.19 |
| chr10_46075820_46076565 | 0.20 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
13747 |
0.25 |
| chrX_19815653_19816006 | 0.20 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chr14_32672268_32672419 | 0.20 |
ENSG00000202337 |
. |
132 |
0.94 |
| chr19_6481304_6482171 | 0.20 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr1_198627689_198627922 | 0.20 |
RP11-553K8.5 |
|
8385 |
0.25 |
| chr5_67577391_67577649 | 0.20 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
1385 |
0.58 |
| chr18_56326442_56326593 | 0.20 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
12101 |
0.14 |
| chrX_17754584_17754752 | 0.20 |
SCML1 |
sex comb on midleg-like 1 (Drosophila) |
920 |
0.7 |
| chr1_143741571_143741722 | 0.20 |
RP6-206I17.4 |
|
21942 |
0.16 |
| chr16_50623254_50623506 | 0.20 |
RP11-401P9.6 |
|
24215 |
0.14 |
| chr14_77838790_77838941 | 0.20 |
SAMD15 |
sterile alpha motif domain containing 15 |
4167 |
0.14 |
| chr2_204572133_204572358 | 0.20 |
CD28 |
CD28 molecule |
829 |
0.71 |
| chr18_9404580_9404814 | 0.20 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
69849 |
0.08 |
| chr14_22947593_22947744 | 0.20 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
2372 |
0.15 |
| chr12_65013240_65013416 | 0.19 |
ENSG00000207546 |
. |
2961 |
0.17 |
| chr1_29254771_29254973 | 0.19 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
13781 |
0.18 |
| chr6_159464766_159465080 | 0.19 |
TAGAP |
T-cell activation RhoGTPase activating protein |
1127 |
0.51 |
| chr12_94805998_94806149 | 0.19 |
CCDC41 |
coiled-coil domain containing 41 |
47649 |
0.14 |
| chr7_115668295_115668446 | 0.18 |
TFEC |
transcription factor EC |
2425 |
0.43 |
| chr1_47726053_47726204 | 0.18 |
STIL |
SCL/TAL1 interrupting locus |
20601 |
0.14 |
| chr7_73509063_73509569 | 0.18 |
LIMK1 |
LIM domain kinase 1 |
1907 |
0.33 |
| chr1_28975271_28975442 | 0.18 |
ENSG00000270103 |
. |
244 |
0.88 |
| chr6_91004568_91004758 | 0.18 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
1798 |
0.43 |
| chr5_75600050_75600201 | 0.18 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr10_173487_173638 | 0.18 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
6843 |
0.24 |
| chr1_236284065_236284436 | 0.18 |
GPR137B |
G protein-coupled receptor 137B |
21582 |
0.19 |
| chr14_22977314_22977663 | 0.18 |
TRAJ15 |
T cell receptor alpha joining 15 |
21092 |
0.09 |
| chr14_22974481_22974956 | 0.18 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18547 |
0.09 |
| chr9_3528863_3529151 | 0.18 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
3003 |
0.37 |
| chr6_28108040_28108408 | 0.17 |
ZKSCAN8 |
zinc finger with KRAB and SCAN domains 8 |
1492 |
0.26 |
| chr3_33478642_33478793 | 0.17 |
UBP1 |
upstream binding protein 1 (LBP-1a) |
2785 |
0.29 |
| chr6_12010262_12011396 | 0.17 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
1404 |
0.51 |
| chr12_92440588_92440837 | 0.17 |
C12orf79 |
chromosome 12 open reading frame 79 |
90085 |
0.08 |
| chr7_37480884_37481045 | 0.17 |
ENSG00000201566 |
. |
6246 |
0.2 |
| chr1_186291714_186291865 | 0.17 |
ENSG00000202025 |
. |
10729 |
0.17 |
| chr13_27234189_27234340 | 0.17 |
WASF3-AS1 |
WASF3 antisense RNA 1 |
18763 |
0.26 |
| chr12_47600587_47601314 | 0.17 |
PCED1B |
PC-esterase domain containing 1B |
9102 |
0.22 |
| chr7_6654683_6655004 | 0.17 |
ZNF853 |
zinc finger protein 853 |
405 |
0.77 |
| chr3_52235228_52235379 | 0.17 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
3143 |
0.15 |
| chr17_21157480_21157631 | 0.17 |
C17orf103 |
chromosome 17 open reading frame 103 |
833 |
0.56 |
| chrX_78196869_78197083 | 0.17 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
3853 |
0.37 |
| chr2_193992778_193992929 | 0.17 |
TMEFF2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
932418 |
0.0 |
| chr7_26141262_26141558 | 0.17 |
ENSG00000266430 |
. |
41451 |
0.15 |
| chrX_118815979_118816490 | 0.17 |
SEPT6 |
septin 6 |
10558 |
0.17 |
| chr13_46745397_46745548 | 0.17 |
ENSG00000240767 |
. |
1589 |
0.32 |
| chr6_52151017_52151168 | 0.16 |
MCM3 |
minichromosome maintenance complex component 3 |
1457 |
0.45 |
| chr11_128589269_128589524 | 0.16 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chr1_197884515_197884666 | 0.16 |
LHX9 |
LIM homeobox 9 |
377 |
0.89 |
| chr14_64195027_64195308 | 0.16 |
SGPP1 |
sphingosine-1-phosphate phosphatase 1 |
410 |
0.84 |
| chr17_29637369_29637894 | 0.16 |
EVI2B |
ecotropic viral integration site 2B |
3471 |
0.16 |
| chr17_29639732_29640055 | 0.16 |
EVI2B |
ecotropic viral integration site 2B |
1209 |
0.36 |
| chr1_111420872_111421052 | 0.16 |
CD53 |
CD53 molecule |
5186 |
0.22 |
| chr7_148543528_148543679 | 0.16 |
ENSG00000251712 |
. |
25499 |
0.15 |
| chr14_99703180_99703331 | 0.16 |
AL109767.1 |
|
26030 |
0.19 |
| chr7_50348449_50348781 | 0.16 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
297 |
0.94 |
| chr11_8228040_8228278 | 0.16 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
37557 |
0.16 |
| chr1_150294067_150294674 | 0.16 |
PRPF3 |
pre-mRNA processing factor 3 |
368 |
0.78 |
| chr4_37686087_37686238 | 0.16 |
RELL1 |
RELT-like 1 |
1836 |
0.39 |
| chr4_144312492_144312643 | 0.16 |
GAB1 |
GRB2-associated binding protein 1 |
96 |
0.98 |
| chr13_43564402_43564714 | 0.16 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
809 |
0.73 |
| chr5_130882612_130883040 | 0.15 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
14100 |
0.29 |
| chr7_107530950_107531101 | 0.15 |
DLD |
dihydrolipoamide dehydrogenase |
390 |
0.88 |
| chr13_97879341_97880219 | 0.15 |
MBNL2 |
muscleblind-like splicing regulator 2 |
5171 |
0.33 |
| chr1_192129351_192129646 | 0.15 |
RGS18 |
regulator of G-protein signaling 18 |
1911 |
0.5 |
| chr3_30657794_30657945 | 0.15 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
9776 |
0.3 |
| chr1_65614810_65615288 | 0.15 |
AK4 |
adenylate kinase 4 |
1163 |
0.58 |
| chr5_43017766_43018004 | 0.15 |
CTD-2035E11.3 |
|
646 |
0.52 |
| chr5_68338922_68339492 | 0.15 |
SLC30A5 |
solute carrier family 30 (zinc transporter), member 5 |
50266 |
0.12 |
| chr3_32280995_32281180 | 0.15 |
CMTM8 |
CKLF-like MARVEL transmembrane domain containing 8 |
916 |
0.41 |
| chr5_35781592_35781782 | 0.15 |
SPEF2 |
sperm flagellar 2 |
2417 |
0.34 |
| chr2_201983400_201983976 | 0.15 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
474 |
0.74 |
| chr4_26887523_26887909 | 0.15 |
STIM2 |
stromal interaction molecule 2 |
24635 |
0.21 |
| chr3_135909805_135909956 | 0.15 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
3516 |
0.32 |
| chr4_39032495_39032784 | 0.15 |
TMEM156 |
transmembrane protein 156 |
1402 |
0.45 |
| chr15_60874237_60874501 | 0.15 |
RORA |
RAR-related orphan receptor A |
10371 |
0.22 |
| chr1_100820231_100820573 | 0.15 |
CDC14A |
cell division cycle 14A |
1897 |
0.33 |
| chr3_151911560_151911983 | 0.15 |
MBNL1 |
muscleblind-like splicing regulator 1 |
74058 |
0.11 |
| chr9_80524074_80524374 | 0.15 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chr12_46062870_46063096 | 0.15 |
ARID2 |
AT rich interactive domain 2 (ARID, RFX-like) |
60465 |
0.15 |
| chr11_65324098_65324348 | 0.15 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
1013 |
0.31 |
| chr6_139466066_139466217 | 0.15 |
HECA |
headcase homolog (Drosophila) |
9892 |
0.26 |
| chr3_98274758_98274909 | 0.15 |
GPR15 |
G protein-coupled receptor 15 |
24090 |
0.13 |
| chr6_91123305_91123789 | 0.15 |
ENSG00000252676 |
. |
32944 |
0.21 |
| chr2_113546245_113546396 | 0.15 |
IL1A |
interleukin 1, alpha |
4153 |
0.2 |
| chr12_12874334_12874808 | 0.15 |
RP11-180M15.4 |
|
2374 |
0.2 |
| chr12_11802043_11802302 | 0.15 |
ETV6 |
ets variant 6 |
616 |
0.81 |
| chr1_11715091_11715564 | 0.15 |
FBXO44 |
F-box protein 44 |
360 |
0.57 |
| chr9_123686452_123686718 | 0.15 |
TRAF1 |
TNF receptor-associated factor 1 |
4462 |
0.25 |
| chr11_2322356_2322739 | 0.14 |
C11orf21 |
chromosome 11 open reading frame 21 |
596 |
0.44 |
| chr13_46755198_46755455 | 0.14 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
1133 |
0.45 |
| chr14_89817609_89817760 | 0.14 |
RP11-356K23.1 |
|
1038 |
0.5 |
| chr14_77873229_77873401 | 0.14 |
FKSG61 |
|
9426 |
0.11 |
| chr2_171555186_171555337 | 0.14 |
AC007277.3 |
|
3377 |
0.22 |
| chr15_44120227_44120469 | 0.14 |
WDR76 |
WD repeat domain 76 |
1162 |
0.29 |
| chr1_112018691_112018961 | 0.14 |
C1orf162 |
chromosome 1 open reading frame 162 |
2335 |
0.16 |
| chrX_19813800_19813951 | 0.14 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
3994 |
0.35 |
| chr5_79550167_79550318 | 0.14 |
SERINC5 |
serine incorporator 5 |
1596 |
0.37 |
| chr3_108541842_108541993 | 0.14 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
298 |
0.94 |
| chr10_112632655_112633305 | 0.14 |
PDCD4-AS1 |
PDCD4 antisense RNA 1 |
989 |
0.38 |
| chr5_98362961_98363596 | 0.14 |
ENSG00000200351 |
. |
90827 |
0.09 |
| chr4_40201423_40201705 | 0.14 |
RHOH |
ras homolog family member H |
400 |
0.87 |
| chr7_37488846_37488997 | 0.14 |
ELMO1 |
engulfment and cell motility 1 |
69 |
0.98 |
| chr6_26198173_26198564 | 0.14 |
HIST1H3D |
histone cluster 1, H3d |
890 |
0.22 |
| chr5_171560548_171560699 | 0.14 |
ENSG00000266671 |
. |
52643 |
0.12 |
| chr1_162843054_162843205 | 0.14 |
C1orf110 |
chromosome 1 open reading frame 110 |
4524 |
0.29 |
| chr11_75063940_75064091 | 0.14 |
ARRB1 |
arrestin, beta 1 |
1142 |
0.41 |
| chr1_231748956_231749347 | 0.14 |
LINC00582 |
long intergenic non-protein coding RNA 582 |
1315 |
0.46 |
| chrX_78403286_78403508 | 0.14 |
GPR174 |
G protein-coupled receptor 174 |
23072 |
0.28 |
| chr14_70111468_70111619 | 0.14 |
KIAA0247 |
KIAA0247 |
33230 |
0.18 |
| chr1_11335043_11335194 | 0.14 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
1127 |
0.47 |
| chrY_1551045_1551310 | 0.14 |
NA |
NA |
> 106 |
NA |
| chr1_100852790_100852941 | 0.14 |
ENSG00000216067 |
. |
8534 |
0.21 |
| chr6_34624121_34624272 | 0.14 |
C6orf106 |
chromosome 6 open reading frame 106 |
15537 |
0.14 |
| chr11_14097018_14097169 | 0.14 |
ENSG00000212365 |
. |
59439 |
0.15 |
| chr3_152019403_152019901 | 0.14 |
MBNL1 |
muscleblind-like splicing regulator 1 |
1665 |
0.42 |
| chrX_19823659_19823810 | 0.14 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
5865 |
0.31 |
| chr5_118606885_118607036 | 0.14 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2511 |
0.28 |
| chrX_1600809_1601304 | 0.13 |
ASMTL |
acetylserotonin O-methyltransferase-like |
28401 |
0.16 |
| chr5_35861092_35861243 | 0.13 |
IL7R |
interleukin 7 receptor |
4173 |
0.24 |
| chr3_187676320_187676496 | 0.13 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
194664 |
0.03 |
| chr21_34760648_34760799 | 0.13 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
14479 |
0.17 |
| chr11_67037576_67038089 | 0.13 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
3880 |
0.14 |
| chr12_1399393_1399544 | 0.13 |
RP5-951N9.2 |
|
95531 |
0.08 |
| chr1_111215065_111215351 | 0.13 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
2447 |
0.28 |
| chr17_44849363_44850053 | 0.13 |
NSF |
N-ethylmaleimide-sensitive factor |
45786 |
0.13 |
| chr1_39500021_39500217 | 0.13 |
NDUFS5 |
NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) |
8129 |
0.18 |
| chr11_57794441_57794592 | 0.13 |
ENSG00000200817 |
. |
232 |
0.91 |
| chr3_119182906_119183082 | 0.13 |
TMEM39A |
transmembrane protein 39A |
465 |
0.79 |
| chr1_29253817_29254129 | 0.13 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12882 |
0.18 |
| chr3_101568772_101569567 | 0.13 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
62 |
0.98 |
| chr10_63813819_63813970 | 0.13 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
4924 |
0.3 |
| chr13_75898854_75899075 | 0.13 |
TBC1D4 |
TBC1 domain family, member 4 |
16703 |
0.24 |
| chr3_151963607_151963763 | 0.13 |
MBNL1 |
muscleblind-like splicing regulator 1 |
22144 |
0.21 |
| chr3_69129681_69129838 | 0.13 |
UBA3 |
ubiquitin-like modifier activating enzyme 3 |
200 |
0.92 |
| chr21_36398548_36398777 | 0.13 |
RUNX1 |
runt-related transcription factor 1 |
22800 |
0.28 |
| chr5_75826683_75826834 | 0.13 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
16476 |
0.24 |
| chr5_148931227_148931707 | 0.13 |
ARHGEF37 |
Rho guanine nucleotide exchange factor (GEF) 37 |
43 |
0.88 |
| chr7_155326313_155326555 | 0.13 |
CNPY1 |
canopy FGF signaling regulator 1 |
99 |
0.97 |
| chr3_45732221_45732372 | 0.13 |
SACM1L |
SAC1 suppressor of actin mutations 1-like (yeast) |
1345 |
0.36 |
| chr9_20242645_20242902 | 0.13 |
ENSG00000221744 |
. |
52229 |
0.16 |
| chr4_39045572_39046195 | 0.13 |
KLHL5 |
kelch-like family member 5 |
776 |
0.67 |
| chr14_91863541_91863979 | 0.13 |
CCDC88C |
coiled-coil domain containing 88C |
19930 |
0.21 |
| chr2_98698576_98698727 | 0.13 |
VWA3B |
von Willebrand factor A domain containing 3B |
4944 |
0.3 |
| chr10_91461400_91461679 | 0.13 |
KIF20B |
kinesin family member 20B |
103 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.3 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.2 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.3 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.0 | GO:2000380 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.0 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.0 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.0 | REACTOME SIGNAL AMPLIFICATION | Genes involved in Signal amplification |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |