Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
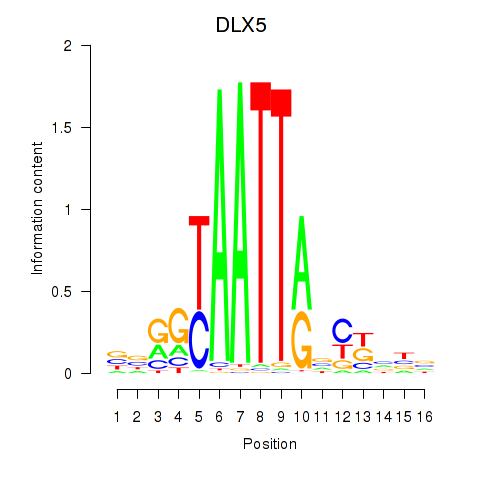
Results for DLX5
Z-value: 0.79
Transcription factors associated with DLX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX5
|
ENSG00000105880.4 | distal-less homeobox 5 |
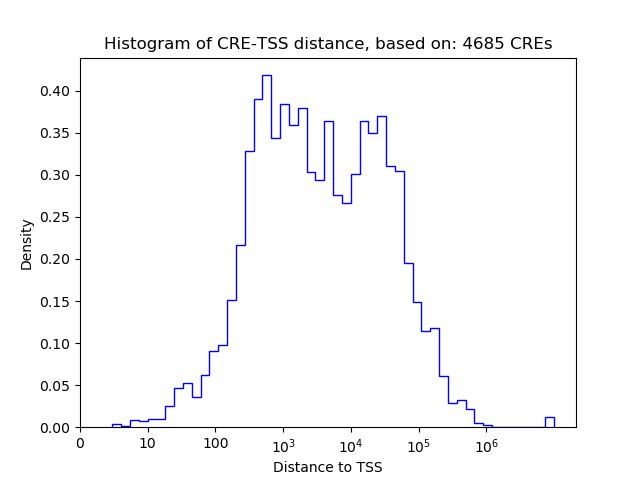
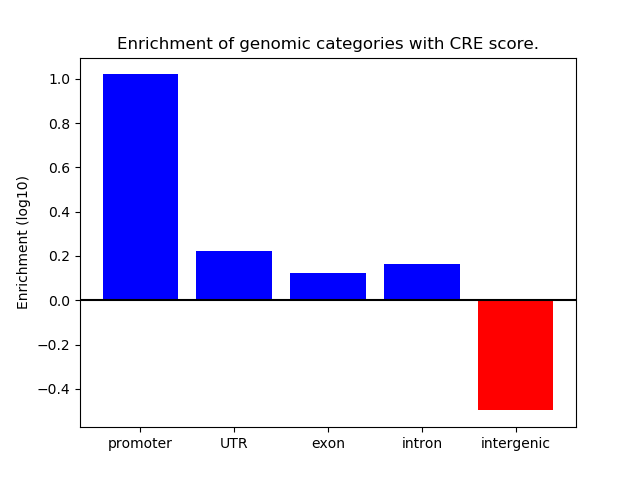
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_96654173_96654357 | DLX5 | 3 | 0.973536 | -0.69 | 3.8e-02 | Click! |
| chr7_96653818_96654015 | DLX5 | 346 | 0.856351 | -0.59 | 9.7e-02 | Click! |
| chr7_96653221_96653741 | DLX5 | 781 | 0.596408 | -0.50 | 1.7e-01 | Click! |
| chr7_96656161_96656312 | DLX5 | 1827 | 0.303265 | 0.46 | 2.1e-01 | Click! |
| chr7_96656340_96657076 | DLX5 | 2299 | 0.256525 | 0.43 | 2.5e-01 | Click! |
Activity of the DLX5 motif across conditions
Conditions sorted by the z-value of the DLX5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
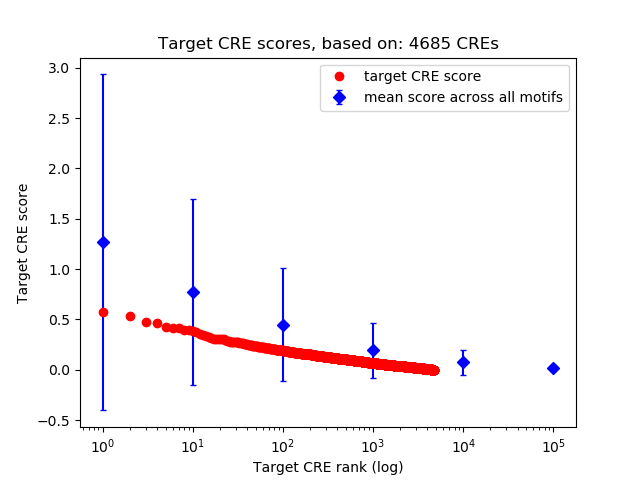
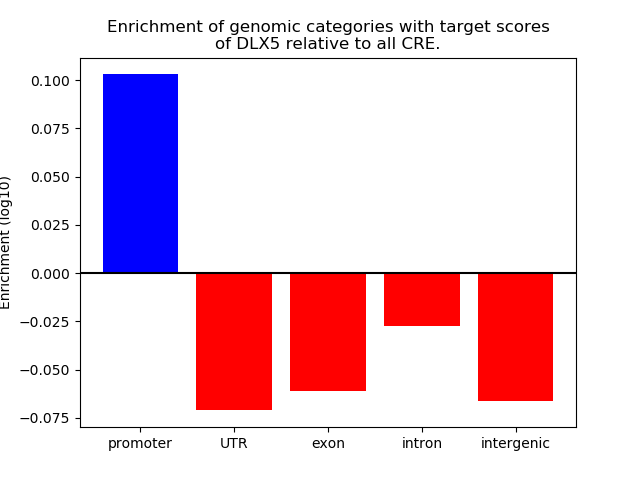
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_26141262_26141558 | 0.58 |
ENSG00000266430 |
. |
41451 |
0.15 |
| chr1_66799510_66799957 | 0.54 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
1861 |
0.5 |
| chrX_19815653_19816006 | 0.47 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chrX_78400540_78401325 | 0.46 |
GPR174 |
G protein-coupled receptor 174 |
25537 |
0.27 |
| chr1_112018691_112018961 | 0.43 |
C1orf162 |
chromosome 1 open reading frame 162 |
2335 |
0.16 |
| chr1_9713063_9713437 | 0.42 |
C1orf200 |
chromosome 1 open reading frame 200 |
1394 |
0.31 |
| chr21_15917916_15918619 | 0.42 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr1_111741177_111741444 | 0.40 |
DENND2D |
DENN/MADD domain containing 2D |
2001 |
0.2 |
| chr20_56194431_56194643 | 0.39 |
ZBP1 |
Z-DNA binding protein 1 |
913 |
0.65 |
| chr11_65324098_65324348 | 0.39 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
1013 |
0.31 |
| chr1_239883177_239883989 | 0.37 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
740 |
0.59 |
| chr3_108543496_108543820 | 0.35 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
2039 |
0.42 |
| chr15_65587802_65587963 | 0.35 |
ENSG00000199568 |
. |
507 |
0.7 |
| chr6_135518404_135518725 | 0.34 |
MYB-AS1 |
MYB antisense RNA 1 |
1431 |
0.4 |
| chr1_162843054_162843205 | 0.33 |
C1orf110 |
chromosome 1 open reading frame 110 |
4524 |
0.29 |
| chr13_99957828_99957979 | 0.31 |
GPR183 |
G protein-coupled receptor 183 |
1756 |
0.38 |
| chr17_29639732_29640055 | 0.31 |
EVI2B |
ecotropic viral integration site 2B |
1209 |
0.36 |
| chr1_66802476_66802627 | 0.31 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
4679 |
0.34 |
| chr2_62426916_62427067 | 0.31 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
3743 |
0.21 |
| chr12_866633_866784 | 0.31 |
WNK1 |
WNK lysine deficient protein kinase 1 |
3976 |
0.25 |
| chr3_121378552_121379115 | 0.30 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
912 |
0.5 |
| chr5_14560333_14560610 | 0.30 |
FAM105A |
family with sequence similarity 105, member A |
21413 |
0.23 |
| chr4_119143007_119143249 | 0.30 |
ENSG00000269893 |
. |
57217 |
0.16 |
| chr17_4615230_4615381 | 0.29 |
ARRB2 |
arrestin, beta 2 |
1311 |
0.23 |
| chr2_85076559_85076967 | 0.28 |
TRABD2A |
TraB domain containing 2A |
31443 |
0.16 |
| chr5_43017766_43018004 | 0.28 |
CTD-2035E11.3 |
|
646 |
0.52 |
| chr5_110560941_110561277 | 0.28 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1325 |
0.51 |
| chr15_94815577_94815728 | 0.28 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
25778 |
0.28 |
| chr3_186745077_186745229 | 0.28 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
1882 |
0.42 |
| chr18_13216452_13216669 | 0.27 |
RP11-794M8.1 |
|
194 |
0.92 |
| chr17_25622078_25622646 | 0.27 |
WSB1 |
WD repeat and SOCS box containing 1 |
990 |
0.48 |
| chr13_27234189_27234340 | 0.27 |
WASF3-AS1 |
WASF3 antisense RNA 1 |
18763 |
0.26 |
| chr13_21050204_21050392 | 0.27 |
ENSG00000263978 |
. |
42313 |
0.15 |
| chr15_55547653_55548107 | 0.27 |
RAB27A |
RAB27A, member RAS oncogene family |
6647 |
0.22 |
| chr11_14097018_14097169 | 0.26 |
ENSG00000212365 |
. |
59439 |
0.15 |
| chr2_47537091_47537242 | 0.26 |
EPCAM |
epithelial cell adhesion molecule |
35131 |
0.15 |
| chr1_53068469_53068939 | 0.26 |
GPX7 |
glutathione peroxidase 7 |
660 |
0.7 |
| chr10_11217828_11218013 | 0.26 |
RP3-323N1.2 |
|
4581 |
0.25 |
| chr5_60626384_60626783 | 0.26 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
1517 |
0.55 |
| chrX_8150685_8150836 | 0.25 |
VCX2 |
variable charge, X-linked 2 |
11452 |
0.28 |
| chrX_30594869_30596024 | 0.25 |
CXorf21 |
chromosome X open reading frame 21 |
515 |
0.84 |
| chr12_75877051_75877218 | 0.25 |
GLIPR1 |
GLI pathogenesis-related 1 |
2150 |
0.29 |
| chr11_122597673_122598179 | 0.25 |
ENSG00000239079 |
. |
907 |
0.67 |
| chr12_75875359_75875792 | 0.25 |
GLIPR1 |
GLI pathogenesis-related 1 |
591 |
0.74 |
| chr17_40400288_40400799 | 0.24 |
RP11-358B23.5 |
|
20062 |
0.1 |
| chr9_95730621_95730837 | 0.24 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
4486 |
0.25 |
| chr10_126745966_126746179 | 0.24 |
ENSG00000264572 |
. |
24633 |
0.19 |
| chr12_120976273_120976798 | 0.24 |
RNF10 |
ring finger protein 10 |
3306 |
0.14 |
| chr15_100881986_100882218 | 0.24 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
108 |
0.98 |
| chr20_8349746_8349924 | 0.23 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
120463 |
0.06 |
| chr18_9404580_9404814 | 0.23 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
69849 |
0.08 |
| chr12_104854519_104854691 | 0.23 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
3826 |
0.33 |
| chr7_150436038_150436258 | 0.23 |
GIMAP5 |
GTPase, IMAP family member 5 |
1712 |
0.32 |
| chr11_128343021_128343263 | 0.23 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
32147 |
0.21 |
| chr19_50062782_50062933 | 0.23 |
NOSIP |
nitric oxide synthase interacting protein |
1030 |
0.27 |
| chr1_47726053_47726204 | 0.23 |
STIL |
SCL/TAL1 interrupting locus |
20601 |
0.14 |
| chr9_131453430_131453735 | 0.23 |
SET |
SET nuclear oncogene |
1314 |
0.27 |
| chr3_119880714_119880865 | 0.22 |
ENSG00000244139 |
. |
40033 |
0.13 |
| chr14_77873229_77873401 | 0.22 |
FKSG61 |
|
9426 |
0.11 |
| chr3_111263576_111263727 | 0.22 |
CD96 |
CD96 molecule |
2654 |
0.36 |
| chr2_25500266_25500584 | 0.22 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
25245 |
0.19 |
| chr5_156476977_156477197 | 0.22 |
HAVCR1 |
hepatitis A virus cellular receptor 1 |
8320 |
0.2 |
| chr11_2322356_2322739 | 0.22 |
C11orf21 |
chromosome 11 open reading frame 21 |
596 |
0.44 |
| chr4_175041157_175041343 | 0.22 |
FBXO8 |
F-box protein 8 |
163564 |
0.04 |
| chr3_63954713_63955178 | 0.22 |
ATXN7 |
ataxin 7 |
1525 |
0.34 |
| chrX_40440586_40440827 | 0.22 |
ATP6AP2 |
ATPase, H+ transporting, lysosomal accessory protein 2 |
443 |
0.86 |
| chr2_44133691_44133842 | 0.22 |
LRPPRC |
leucine-rich pentatricopeptide repeat containing |
7343 |
0.2 |
| chr8_66742389_66742540 | 0.21 |
PDE7A |
phosphodiesterase 7A |
8519 |
0.29 |
| chr6_32158116_32158500 | 0.21 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
345 |
0.7 |
| chr11_75063727_75063930 | 0.21 |
ARRB1 |
arrestin, beta 1 |
955 |
0.48 |
| chr8_39797291_39797650 | 0.21 |
IDO2 |
indoleamine 2,3-dioxygenase 2 |
4996 |
0.18 |
| chr6_41650642_41650943 | 0.21 |
AL035588.1 |
|
3172 |
0.18 |
| chr1_204482801_204482952 | 0.21 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
2635 |
0.26 |
| chr12_11803298_11803449 | 0.21 |
ETV6 |
ets variant 6 |
585 |
0.83 |
| chr2_25473265_25473416 | 0.21 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
1840 |
0.39 |
| chr11_14995196_14995439 | 0.21 |
CALCA |
calcitonin-related polypeptide alpha |
1417 |
0.53 |
| chr18_66381478_66381997 | 0.21 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
557 |
0.55 |
| chr10_75935885_75936201 | 0.21 |
ADK |
adenosine kinase |
401 |
0.88 |
| chr1_179856871_179857022 | 0.20 |
TOR1AIP1 |
torsin A interacting protein 1 |
4922 |
0.16 |
| chr8_102802981_102803132 | 0.20 |
NCALD |
neurocalcin delta |
119 |
0.98 |
| chr15_75080868_75081404 | 0.20 |
ENSG00000264386 |
. |
38 |
0.76 |
| chr4_109056842_109056993 | 0.20 |
LEF1 |
lymphoid enhancer-binding factor 1 |
30540 |
0.19 |
| chr12_11802043_11802302 | 0.20 |
ETV6 |
ets variant 6 |
616 |
0.81 |
| chr13_41555714_41555943 | 0.20 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
590 |
0.77 |
| chr15_57210828_57211163 | 0.20 |
TCF12 |
transcription factor 12 |
35 |
0.75 |
| chr6_126068689_126068849 | 0.20 |
HEY2 |
hes-related family bHLH transcription factor with YRPW motif 2 |
41 |
0.97 |
| chr4_84035334_84035716 | 0.20 |
PLAC8 |
placenta-specific 8 |
343 |
0.91 |
| chr14_22974481_22974956 | 0.20 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18547 |
0.09 |
| chr5_118606012_118606179 | 0.20 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
1646 |
0.37 |
| chr1_27115315_27115664 | 0.20 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
70 |
0.96 |
| chr6_107154119_107154270 | 0.20 |
ENSG00000202285 |
. |
32735 |
0.17 |
| chr2_88124610_88124761 | 0.20 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
601 |
0.82 |
| chr22_41033042_41033281 | 0.20 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
466 |
0.84 |
| chr1_153918448_153918692 | 0.20 |
DENND4B |
DENN/MADD domain containing 4B |
602 |
0.54 |
| chr1_214484548_214484699 | 0.20 |
SMYD2 |
SET and MYND domain containing 2 |
19697 |
0.24 |
| chr15_61013382_61013533 | 0.19 |
ENSG00000212625 |
. |
15511 |
0.2 |
| chr1_169676400_169676585 | 0.19 |
SELL |
selectin L |
4347 |
0.23 |
| chr12_51818248_51818602 | 0.19 |
RP11-607P23.1 |
|
30 |
0.62 |
| chr17_75456326_75456913 | 0.19 |
SEPT9 |
septin 9 |
4171 |
0.18 |
| chr13_41164420_41164571 | 0.19 |
AL133318.1 |
Uncharacterized protein |
53172 |
0.14 |
| chr19_46194641_46194820 | 0.19 |
SNRPD2 |
small nuclear ribonucleoprotein D2 polypeptide 16.5kDa |
299 |
0.72 |
| chr22_40858929_40859404 | 0.19 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
256 |
0.92 |
| chr3_136537398_136537875 | 0.19 |
SLC35G2 |
solute carrier family 35, member G2 |
225 |
0.92 |
| chr17_45725211_45725362 | 0.19 |
RP11-580I16.2 |
|
1482 |
0.26 |
| chr6_24934739_24934890 | 0.19 |
FAM65B |
family with sequence similarity 65, member B |
1374 |
0.5 |
| chr5_169708113_169708278 | 0.19 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
13864 |
0.22 |
| chr14_22447473_22447624 | 0.19 |
ENSG00000238634 |
. |
163339 |
0.03 |
| chr1_65096460_65096611 | 0.19 |
ENSG00000264470 |
. |
51005 |
0.17 |
| chr6_2222723_2222882 | 0.19 |
GMDS |
GDP-mannose 4,6-dehydratase |
23124 |
0.28 |
| chr1_150737696_150737976 | 0.19 |
CTSS |
cathepsin S |
432 |
0.77 |
| chr11_61158553_61158704 | 0.18 |
TMEM216 |
transmembrane protein 216 |
531 |
0.65 |
| chr2_161995257_161995554 | 0.18 |
TANK |
TRAF family member-associated NFKB activator |
1939 |
0.4 |
| chr17_81147907_81148162 | 0.18 |
METRNL |
meteorin, glial cell differentiation regulator-like |
96040 |
0.08 |
| chr11_60928486_60928645 | 0.18 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
43 |
0.97 |
| chr1_244999063_244999557 | 0.18 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
392 |
0.85 |
| chr10_12238316_12238700 | 0.18 |
CDC123 |
cell division cycle 123 |
277 |
0.57 |
| chrX_64196083_64196443 | 0.18 |
ZC4H2 |
zinc finger, C4H2 domain containing |
65 |
0.99 |
| chr22_22121341_22121492 | 0.18 |
ENSG00000200985 |
. |
25307 |
0.12 |
| chrX_54557248_54557562 | 0.18 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
730 |
0.73 |
| chr3_12228694_12228845 | 0.18 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
27918 |
0.22 |
| chr17_1811216_1811594 | 0.18 |
RPA1 |
replication protein A1, 70kDa |
29076 |
0.13 |
| chrX_78200973_78201449 | 0.18 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
293 |
0.95 |
| chr4_39979745_39980089 | 0.18 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
341 |
0.9 |
| chr5_58883529_58883730 | 0.18 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
1304 |
0.62 |
| chr8_128988445_128988635 | 0.18 |
ENSG00000221771 |
. |
15661 |
0.16 |
| chr21_45341679_45341830 | 0.17 |
ENSG00000199598 |
. |
1972 |
0.29 |
| chrX_78202204_78202379 | 0.17 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
1373 |
0.61 |
| chr1_79322114_79322265 | 0.17 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
61381 |
0.15 |
| chr16_23265949_23266165 | 0.17 |
CTC-391G2.1 |
|
41632 |
0.14 |
| chr12_131796680_131796831 | 0.17 |
ENSG00000212251 |
. |
12459 |
0.23 |
| chr9_130823963_130824114 | 0.17 |
RP11-379C10.1 |
|
4397 |
0.11 |
| chr14_31879526_31879677 | 0.17 |
HEATR5A |
HEAT repeat containing 5A |
7420 |
0.17 |
| chr19_13859245_13859441 | 0.17 |
CCDC130 |
coiled-coil domain containing 130 |
632 |
0.57 |
| chr1_183843426_183843577 | 0.17 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
69211 |
0.11 |
| chr7_150267315_150267508 | 0.17 |
GIMAP4 |
GTPase, IMAP family member 4 |
2887 |
0.27 |
| chr6_76203613_76203764 | 0.17 |
FILIP1 |
filamin A interacting protein 1 |
234 |
0.93 |
| chrX_118751905_118752056 | 0.17 |
NKRF |
NFKB repressing factor |
12122 |
0.15 |
| chr6_2854275_2854463 | 0.17 |
ENSG00000266750 |
. |
28 |
0.98 |
| chr3_30673309_30673499 | 0.17 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
25311 |
0.24 |
| chr20_43599792_43600332 | 0.17 |
STK4 |
serine/threonine kinase 4 |
4895 |
0.16 |
| chr1_206646546_206646697 | 0.17 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
2783 |
0.21 |
| chr1_51442241_51442392 | 0.17 |
CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
6700 |
0.22 |
| chr20_3388735_3389323 | 0.17 |
C20orf194 |
chromosome 20 open reading frame 194 |
757 |
0.58 |
| chr3_64326564_64326736 | 0.17 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
72995 |
0.11 |
| chr13_75898854_75899075 | 0.17 |
TBC1D4 |
TBC1 domain family, member 4 |
16703 |
0.24 |
| chr8_82597903_82598285 | 0.17 |
IMPA1 |
inositol(myo)-1(or 4)-monophosphatase 1 |
27 |
0.97 |
| chr21_36308793_36309139 | 0.17 |
RUNX1 |
runt-related transcription factor 1 |
46879 |
0.19 |
| chr14_62170526_62170677 | 0.17 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
6261 |
0.26 |
| chr15_60874237_60874501 | 0.17 |
RORA |
RAR-related orphan receptor A |
10371 |
0.22 |
| chr7_145332771_145332922 | 0.16 |
ENSG00000264018 |
. |
460334 |
0.01 |
| chr3_152878700_152878913 | 0.16 |
RP11-529G21.2 |
|
758 |
0.56 |
| chr17_29359555_29359720 | 0.16 |
RP11-271K11.5 |
|
18407 |
0.11 |
| chrX_102508933_102509363 | 0.16 |
TCEAL8 |
transcription elongation factor A (SII)-like 8 |
965 |
0.51 |
| chr12_12874334_12874808 | 0.16 |
RP11-180M15.4 |
|
2374 |
0.2 |
| chr9_100565871_100566193 | 0.16 |
FOXE1 |
forkhead box E1 (thyroid transcription factor 2) |
49504 |
0.13 |
| chr10_30756907_30757058 | 0.16 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
29231 |
0.19 |
| chr9_120468751_120468935 | 0.16 |
ENSG00000201444 |
. |
1537 |
0.37 |
| chr3_128950544_128950778 | 0.16 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
17788 |
0.14 |
| chr7_6522650_6523366 | 0.16 |
FLJ20306 |
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
86 |
0.87 |
| chr17_80817564_80818630 | 0.16 |
TBCD |
tubulin folding cofactor D |
173 |
0.94 |
| chr6_26204271_26204655 | 0.16 |
HIST1H4E |
histone cluster 1, H4e |
395 |
0.59 |
| chr9_88897597_88898295 | 0.16 |
ISCA1 |
iron-sulfur cluster assembly 1 |
270 |
0.91 |
| chr4_183525693_183525844 | 0.16 |
ENSG00000251714 |
. |
27073 |
0.21 |
| chr1_162467642_162468629 | 0.16 |
UHMK1 |
U2AF homology motif (UHM) kinase 1 |
502 |
0.8 |
| chr9_75766606_75766757 | 0.16 |
ANXA1 |
annexin A1 |
8 |
0.99 |
| chr9_36573868_36574019 | 0.16 |
MELK |
maternal embryonic leucine zipper kinase |
1035 |
0.67 |
| chr8_16211740_16211891 | 0.16 |
MSR1 |
macrophage scavenger receptor 1 |
139232 |
0.05 |
| chr8_74283688_74283839 | 0.16 |
RP11-434I12.2 |
|
15067 |
0.24 |
| chr15_31634841_31635126 | 0.16 |
KLF13 |
Kruppel-like factor 13 |
2847 |
0.4 |
| chr1_245215084_245215235 | 0.16 |
ENSG00000251754 |
. |
7929 |
0.17 |
| chr10_47640402_47640757 | 0.16 |
ANTXRLP1 |
anthrax toxin receptor-like pseudogene 1 |
4212 |
0.25 |
| chr1_89992055_89992206 | 0.16 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
1668 |
0.45 |
| chr13_74818552_74818703 | 0.16 |
ENSG00000206617 |
. |
44724 |
0.19 |
| chr11_122040052_122040217 | 0.16 |
ENSG00000207994 |
. |
17118 |
0.17 |
| chr8_66843866_66844135 | 0.16 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
89795 |
0.08 |
| chrX_150564855_150565441 | 0.16 |
VMA21 |
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
110 |
0.98 |
| chr3_57162010_57162273 | 0.16 |
IL17RD |
interleukin 17 receptor D |
14378 |
0.18 |
| chr8_8204138_8204819 | 0.16 |
SGK223 |
Tyrosine-protein kinase SgK223 |
34779 |
0.19 |
| chr7_135656834_135656985 | 0.16 |
MTPN |
myotrophin |
5155 |
0.19 |
| chr20_36040358_36040543 | 0.16 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
27897 |
0.21 |
| chr3_18478950_18479101 | 0.16 |
SATB1 |
SATB homeobox 1 |
1159 |
0.53 |
| chr6_136824219_136824370 | 0.16 |
MAP7 |
microtubule-associated protein 7 |
22827 |
0.18 |
| chr1_27463127_27463278 | 0.16 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
17771 |
0.19 |
| chr13_31310422_31310683 | 0.16 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
907 |
0.71 |
| chr7_102531578_102531729 | 0.16 |
LRRC17 |
leucine rich repeat containing 17 |
21785 |
0.16 |
| chr1_247553346_247553582 | 0.16 |
NLRP3 |
NLR family, pyrin domain containing 3 |
25994 |
0.15 |
| chr1_2478602_2479100 | 0.16 |
TNFRSF14 |
tumor necrosis factor receptor superfamily, member 14 |
8227 |
0.09 |
| chr8_134081639_134081790 | 0.15 |
SLA |
Src-like-adaptor |
9111 |
0.25 |
| chr5_68338922_68339492 | 0.15 |
SLC30A5 |
solute carrier family 30 (zinc transporter), member 5 |
50266 |
0.12 |
| chr1_64977342_64977493 | 0.15 |
CACHD1 |
cache domain containing 1 |
40942 |
0.18 |
| chr3_71349536_71349687 | 0.15 |
FOXP1 |
forkhead box P1 |
4300 |
0.28 |
| chr20_36543212_36543480 | 0.15 |
VSTM2L |
V-set and transmembrane domain containing 2 like |
11802 |
0.23 |
| chr12_2754166_2754317 | 0.15 |
CACNA1C-AS2 |
CACNA1C antisense RNA 2 |
27145 |
0.16 |
| chr5_75600050_75600201 | 0.15 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr10_126835424_126835575 | 0.15 |
CTBP2 |
C-terminal binding protein 2 |
11786 |
0.28 |
| chr18_60982497_60982692 | 0.15 |
RP11-28F1.2 |
|
1279 |
0.41 |
| chr15_60420104_60420458 | 0.15 |
FOXB1 |
forkhead box B1 |
123860 |
0.06 |
| chr4_154411300_154411567 | 0.15 |
KIAA0922 |
KIAA0922 |
23932 |
0.22 |
| chr3_53288776_53289025 | 0.15 |
TKT |
transketolase |
1116 |
0.42 |
| chr6_46097188_46097569 | 0.15 |
RP1-8B1.4 |
|
50 |
0.84 |
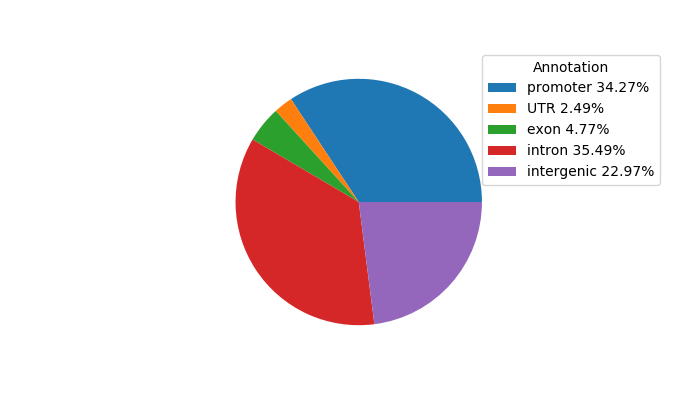
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.3 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.1 | 0.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.2 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.1 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.2 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.2 | GO:0052033 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.3 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.1 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:2000351 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:2000380 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.0 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0045357 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:1904936 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:1901663 | quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.7 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.2 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |