Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
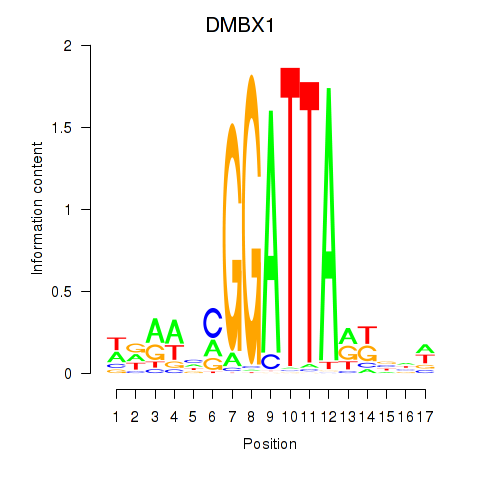
Results for DMBX1
Z-value: 0.45
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.6 | diencephalon/mesencephalon homeobox 1 |
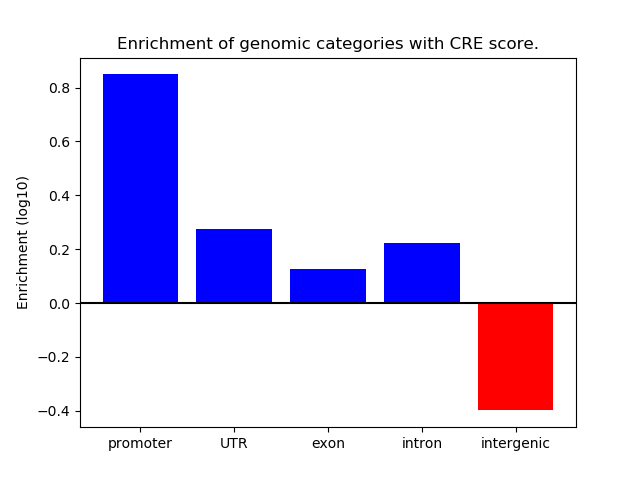
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_46955981_46956132 | DMBX1 | 16612 | 0.157429 | 0.54 | 1.4e-01 | Click! |
| chr1_46982968_46983119 | DMBX1 | 10374 | 0.151850 | -0.26 | 5.0e-01 | Click! |
| chr1_46988200_46988370 | DMBX1 | 15616 | 0.138486 | -0.24 | 5.4e-01 | Click! |
| chr1_46987836_46987987 | DMBX1 | 15242 | 0.139395 | 0.20 | 6.0e-01 | Click! |
| chr1_46955046_46955364 | DMBX1 | 17463 | 0.156152 | 0.14 | 7.2e-01 | Click! |
Activity of the DMBX1 motif across conditions
Conditions sorted by the z-value of the DMBX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
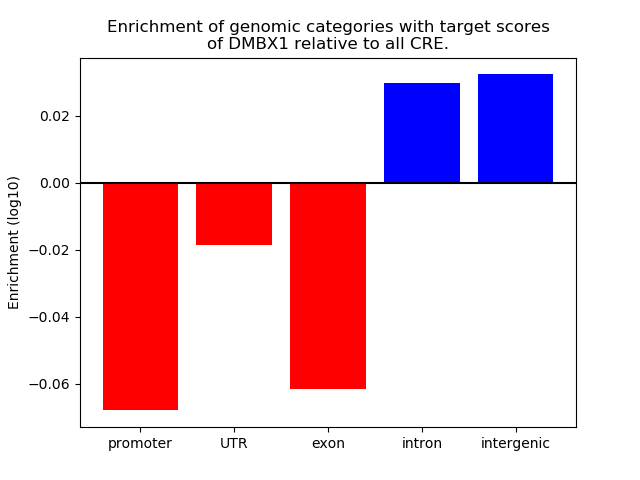
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_5835221_5835372 | 0.27 |
SOX11 |
SRY (sex determining region Y)-box 11 |
2497 |
0.27 |
| chr16_75327785_75327936 | 0.19 |
ENSG00000252101 |
. |
7164 |
0.15 |
| chr5_98111445_98111873 | 0.18 |
RGMB |
repulsive guidance molecule family member b |
2320 |
0.3 |
| chrX_34671991_34672311 | 0.17 |
TMEM47 |
transmembrane protein 47 |
3254 |
0.41 |
| chr13_49393197_49393348 | 0.16 |
ENSG00000222391 |
. |
66980 |
0.12 |
| chr12_19335217_19335368 | 0.16 |
ENSG00000240993 |
. |
5769 |
0.23 |
| chr6_134211072_134211606 | 0.15 |
TCF21 |
transcription factor 21 |
866 |
0.5 |
| chr7_19408295_19408500 | 0.14 |
FERD3L |
Fer3-like bHLH transcription factor |
223353 |
0.02 |
| chr10_70321671_70321944 | 0.14 |
TET1 |
tet methylcytosine dioxygenase 1 |
1394 |
0.42 |
| chr10_114658260_114658457 | 0.14 |
RP11-57H14.3 |
|
9864 |
0.23 |
| chr2_201473941_201474433 | 0.14 |
AOX1 |
aldehyde oxidase 1 |
21290 |
0.2 |
| chr12_45266809_45266960 | 0.13 |
NELL2 |
NEL-like 2 (chicken) |
2367 |
0.35 |
| chr8_17579539_17579690 | 0.13 |
MTUS1 |
microtubule associated tumor suppressor 1 |
116 |
0.96 |
| chr15_35949903_35950054 | 0.13 |
DPH6 |
diphthamine biosynthesis 6 |
111585 |
0.07 |
| chr10_102806768_102807199 | 0.13 |
KAZALD1 |
Kazal-type serine peptidase inhibitor domain 1 |
14615 |
0.1 |
| chr2_36585229_36585663 | 0.13 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
1832 |
0.5 |
| chr6_20876734_20876885 | 0.12 |
RP3-348I23.2 |
|
75884 |
0.12 |
| chr7_116661278_116661429 | 0.12 |
ST7 |
suppression of tumorigenicity 7 |
1022 |
0.54 |
| chr15_38545953_38546360 | 0.12 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
774 |
0.79 |
| chr16_55090478_55090815 | 0.12 |
IRX5 |
iroquois homeobox 5 |
124662 |
0.06 |
| chr10_32165838_32165989 | 0.12 |
ARHGAP12 |
Rho GTPase activating protein 12 |
22805 |
0.24 |
| chr8_77806062_77806213 | 0.11 |
ENSG00000266712 |
. |
72949 |
0.12 |
| chr2_11051733_11051998 | 0.11 |
KCNF1 |
potassium voltage-gated channel, subfamily F, member 1 |
198 |
0.95 |
| chr10_22918703_22918854 | 0.11 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
38136 |
0.21 |
| chr2_134325508_134325697 | 0.11 |
NCKAP5 |
NCK-associated protein 5 |
429 |
0.88 |
| chr15_48089071_48089222 | 0.11 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
30369 |
0.24 |
| chr11_125497097_125497248 | 0.11 |
CHEK1 |
checkpoint kinase 1 |
529 |
0.7 |
| chr6_113953461_113953992 | 0.11 |
ENSG00000266650 |
. |
29609 |
0.21 |
| chr4_7937771_7938061 | 0.11 |
AC097381.1 |
|
2812 |
0.26 |
| chrX_37659049_37659200 | 0.11 |
CYBB |
cytochrome b-245, beta polypeptide |
19807 |
0.21 |
| chr22_19701377_19701773 | 0.11 |
SEPT5 |
septin 5 |
412 |
0.83 |
| chr8_49831086_49831370 | 0.11 |
SNAI2 |
snail family zinc finger 2 |
2760 |
0.41 |
| chr18_8707466_8707765 | 0.11 |
RP11-674N23.1 |
|
4 |
0.73 |
| chr4_182878219_182878616 | 0.11 |
ENSG00000251742 |
. |
122156 |
0.06 |
| chr1_54198514_54198700 | 0.10 |
GLIS1 |
GLIS family zinc finger 1 |
1270 |
0.46 |
| chr14_53301814_53302501 | 0.10 |
FERMT2 |
fermitin family member 2 |
29082 |
0.15 |
| chr15_40269217_40269368 | 0.10 |
EIF2AK4 |
eukaryotic translation initiation factor 2 alpha kinase 4 |
23116 |
0.14 |
| chrY_2709555_2710183 | 0.10 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
92 |
0.98 |
| chr12_3308931_3310293 | 0.10 |
TSPAN9 |
tetraspanin 9 |
729 |
0.75 |
| chr18_53537512_53537663 | 0.10 |
TCF4 |
transcription factor 4 |
205569 |
0.03 |
| chr13_73082008_73082159 | 0.10 |
ENSG00000251715 |
. |
47344 |
0.15 |
| chr19_2852298_2852449 | 0.10 |
AC006130.3 |
|
6341 |
0.11 |
| chr5_3598562_3598819 | 0.10 |
CTD-2012M11.3 |
|
1612 |
0.42 |
| chr8_117698220_117698371 | 0.10 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
69728 |
0.11 |
| chr21_17498457_17498608 | 0.10 |
ENSG00000252273 |
. |
90703 |
0.1 |
| chr1_213500454_213500722 | 0.10 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
275984 |
0.01 |
| chr14_75900439_75900590 | 0.10 |
JDP2 |
Jun dimerization protein 2 |
1677 |
0.37 |
| chr20_56800653_56800804 | 0.09 |
ANKRD60 |
ankyrin repeat domain 60 |
2967 |
0.27 |
| chr17_32367186_32367371 | 0.09 |
CTC-304I17.4 |
|
41965 |
0.12 |
| chr2_177027038_177027323 | 0.09 |
HOXD3 |
homeobox D3 |
1561 |
0.19 |
| chr5_9546839_9547056 | 0.09 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
760 |
0.6 |
| chr6_99292512_99292663 | 0.09 |
POU3F2 |
POU class 3 homeobox 2 |
10007 |
0.31 |
| chr15_96878987_96879612 | 0.09 |
ENSG00000222651 |
. |
2809 |
0.21 |
| chr18_12279738_12279916 | 0.09 |
ENSG00000199702 |
. |
24366 |
0.15 |
| chr7_65263493_65263644 | 0.09 |
ENSG00000200670 |
. |
16091 |
0.16 |
| chr7_42275396_42275690 | 0.09 |
GLI3 |
GLI family zinc finger 3 |
1069 |
0.7 |
| chr6_52435936_52436144 | 0.09 |
TRAM2 |
translocation associated membrane protein 2 |
5673 |
0.27 |
| chr7_155166018_155166497 | 0.09 |
BLACE |
B-cell acute lymphoblastic leukemia expressed |
5628 |
0.19 |
| chr10_84887384_84887535 | 0.09 |
ENSG00000200774 |
. |
59632 |
0.17 |
| chr14_73477705_73477954 | 0.09 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
15985 |
0.17 |
| chr7_111384662_111384813 | 0.09 |
DOCK4 |
dedicator of cytokinesis 4 |
39747 |
0.19 |
| chr4_99509312_99509463 | 0.09 |
TSPAN5 |
tetraspanin 5 |
69399 |
0.11 |
| chr4_103741886_103742050 | 0.09 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
4731 |
0.17 |
| chr12_14133878_14134449 | 0.09 |
GRIN2B |
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
1110 |
0.67 |
| chr18_52987383_52987534 | 0.09 |
TCF4 |
transcription factor 4 |
1759 |
0.5 |
| chr3_181433689_181433840 | 0.09 |
SOX2 |
SRY (sex determining region Y)-box 2 |
4042 |
0.27 |
| chr2_216298025_216298259 | 0.09 |
FN1 |
fibronectin 1 |
2648 |
0.29 |
| chr19_47756874_47757099 | 0.09 |
CCDC9 |
coiled-coil domain containing 9 |
2730 |
0.19 |
| chr8_136478593_136478847 | 0.09 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
7770 |
0.25 |
| chr21_18376217_18376955 | 0.08 |
ENSG00000239023 |
. |
285269 |
0.01 |
| chr6_80246556_80246791 | 0.08 |
LCA5 |
Leber congenital amaurosis 5 |
452 |
0.9 |
| chr8_93082068_93082219 | 0.08 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
1472 |
0.57 |
| chr18_57514598_57514749 | 0.08 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
52507 |
0.14 |
| chr4_53580350_53580621 | 0.08 |
ENSG00000212588 |
. |
1069 |
0.38 |
| chr3_21955222_21955373 | 0.08 |
ENSG00000221384 |
. |
84067 |
0.11 |
| chr15_36761589_36761769 | 0.08 |
C15orf41 |
chromosome 15 open reading frame 41 |
110133 |
0.08 |
| chr2_164591967_164592245 | 0.08 |
FIGN |
fidgetin |
411 |
0.92 |
| chr6_132520617_132520826 | 0.08 |
ENSG00000265669 |
. |
84318 |
0.1 |
| chr1_118728319_118728470 | 0.08 |
SPAG17 |
sperm associated antigen 17 |
548 |
0.86 |
| chr20_18686497_18686694 | 0.08 |
RP11-379J5.5 |
|
7242 |
0.23 |
| chr1_95006423_95007262 | 0.08 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
351 |
0.93 |
| chr4_177524189_177524340 | 0.08 |
RP11-313E19.2 |
|
66508 |
0.14 |
| chr10_21811077_21811228 | 0.08 |
SKIDA1 |
SKI/DACH domain containing 1 |
3459 |
0.17 |
| chr11_73849376_73849624 | 0.08 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
32529 |
0.14 |
| chr13_44842345_44842496 | 0.08 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
105558 |
0.06 |
| chr3_55753015_55753166 | 0.08 |
ENSG00000264013 |
. |
133532 |
0.05 |
| chr4_164265165_164265569 | 0.08 |
NPY5R |
neuropeptide Y receptor Y5 |
113 |
0.9 |
| chr4_44918934_44919085 | 0.08 |
GNPDA2 |
glucosamine-6-phosphate deaminase 2 |
190397 |
0.03 |
| chr7_135431242_135431466 | 0.08 |
FAM180A |
family with sequence similarity 180, member A |
2106 |
0.28 |
| chr2_175711229_175711408 | 0.08 |
CHN1 |
chimerin 1 |
175 |
0.97 |
| chr5_54048752_54048965 | 0.08 |
ENSG00000221073 |
. |
99918 |
0.08 |
| chr9_109876632_109876800 | 0.08 |
RP11-508N12.2 |
|
11447 |
0.29 |
| chr6_24283883_24284034 | 0.08 |
KAAG1 |
kidney associated antigen 1 |
73173 |
0.09 |
| chr1_157014491_157014673 | 0.08 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
283 |
0.91 |
| chr8_106415860_106416011 | 0.08 |
ZFPM2 |
zinc finger protein, FOG family member 2 |
45229 |
0.2 |
| chr17_8284687_8284838 | 0.08 |
RPL26 |
ribosomal protein L26 |
865 |
0.39 |
| chr1_153957648_153957888 | 0.08 |
RAB13 |
RAB13, member RAS oncogene family |
1060 |
0.29 |
| chr1_232689998_232690149 | 0.08 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
7231 |
0.32 |
| chr5_150181131_150181300 | 0.08 |
AC010441.1 |
|
23348 |
0.14 |
| chr12_104199013_104199284 | 0.08 |
RP11-650K20.2 |
|
14240 |
0.14 |
| chr3_133020367_133020518 | 0.08 |
RP11-402L6.1 |
|
44410 |
0.17 |
| chr3_79066759_79066910 | 0.08 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
860 |
0.75 |
| chr12_22093968_22094205 | 0.08 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
81 |
0.98 |
| chr13_49686233_49686384 | 0.08 |
FNDC3A |
fibronectin type III domain containing 3A |
1859 |
0.39 |
| chr17_35851874_35852313 | 0.07 |
DUSP14 |
dual specificity phosphatase 14 |
523 |
0.79 |
| chr17_4132905_4133056 | 0.07 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
34162 |
0.11 |
| chr14_85998679_85999082 | 0.07 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
2308 |
0.36 |
| chr3_114477857_114478359 | 0.07 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
10 |
0.99 |
| chr12_49607635_49607786 | 0.07 |
TUBA1C |
tubulin, alpha 1c |
13999 |
0.12 |
| chr16_64412908_64413059 | 0.07 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
680598 |
0.0 |
| chrX_46763305_46763542 | 0.07 |
CXorf31 |
chromosome X open reading frame 31 |
4285 |
0.23 |
| chr4_186694713_186694864 | 0.07 |
SORBS2 |
sorbin and SH3 domain containing 2 |
1642 |
0.46 |
| chr3_98700054_98700344 | 0.07 |
ENSG00000207331 |
. |
72896 |
0.1 |
| chr18_11856994_11857516 | 0.07 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
184 |
0.63 |
| chr9_100069620_100070257 | 0.07 |
CCDC180 |
coiled-coil domain containing 180 |
3 |
0.98 |
| chr3_125931709_125931911 | 0.07 |
ALDH1L1-AS2 |
ALDH1L1 antisense RNA 2 |
5712 |
0.19 |
| chr17_20756313_20756464 | 0.07 |
RP11-344E13.4 |
|
2871 |
0.23 |
| chr7_32626820_32626992 | 0.07 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
44039 |
0.18 |
| chr14_102434123_102434305 | 0.07 |
DYNC1H1 |
dynein, cytoplasmic 1, heavy chain 1 |
3349 |
0.27 |
| chr2_27877972_27878123 | 0.07 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
8291 |
0.11 |
| chrX_21942836_21942987 | 0.07 |
SMS |
spermine synthase |
15780 |
0.2 |
| chr6_166422463_166422818 | 0.07 |
ENSG00000252196 |
. |
66811 |
0.12 |
| chr5_73926204_73926355 | 0.07 |
HEXB |
hexosaminidase B (beta polypeptide) |
9569 |
0.21 |
| chr1_154840943_154841324 | 0.07 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
338 |
0.86 |
| chr6_3844247_3844398 | 0.07 |
FAM50B |
family with sequence similarity 50, member B |
5298 |
0.22 |
| chr14_95402811_95402962 | 0.07 |
GSC |
goosecoid homeobox |
166324 |
0.03 |
| chr15_37403154_37403481 | 0.07 |
MEIS2 |
Meis homeobox 2 |
9813 |
0.23 |
| chr4_7939548_7939699 | 0.07 |
AC097381.1 |
|
1105 |
0.47 |
| chr6_141587373_141587724 | 0.07 |
ENSG00000222764 |
. |
220001 |
0.02 |
| chr7_102554176_102554437 | 0.07 |
LRRC17 |
leucine rich repeat containing 17 |
854 |
0.61 |
| chr12_21656545_21656817 | 0.07 |
GOLT1B |
golgi transport 1B |
1932 |
0.26 |
| chr13_45147711_45147927 | 0.07 |
TSC22D1 |
TSC22 domain family, member 1 |
2573 |
0.38 |
| chr11_115372100_115372323 | 0.07 |
CADM1 |
cell adhesion molecule 1 |
2896 |
0.39 |
| chr12_3807804_3807975 | 0.07 |
ENSG00000222338 |
. |
27604 |
0.21 |
| chr5_112606936_112607087 | 0.07 |
MCC |
mutated in colorectal cancers |
23635 |
0.23 |
| chr6_26575872_26576023 | 0.07 |
ABT1 |
activator of basal transcription 1 |
21233 |
0.14 |
| chr13_48477961_48478112 | 0.07 |
SUCLA2 |
succinate-CoA ligase, ADP-forming, beta subunit |
97074 |
0.08 |
| chr3_76360042_76360193 | 0.07 |
ENSG00000212104 |
. |
107014 |
0.08 |
| chr2_219828438_219828589 | 0.07 |
AC097468.7 |
|
1888 |
0.18 |
| chr1_78961164_78961362 | 0.07 |
PTGFR |
prostaglandin F receptor (FP) |
4506 |
0.32 |
| chr16_59669437_59669588 | 0.07 |
ENSG00000200062 |
. |
95482 |
0.09 |
| chr12_42979856_42980080 | 0.07 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
3510 |
0.33 |
| chr21_17443239_17443390 | 0.07 |
ENSG00000252273 |
. |
35485 |
0.23 |
| chr6_8100517_8100668 | 0.07 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
1687 |
0.45 |
| chr7_127744609_127744845 | 0.07 |
ENSG00000207588 |
. |
22814 |
0.21 |
| chr10_114552786_114553214 | 0.07 |
RP11-57H14.4 |
|
30249 |
0.21 |
| chr17_7493877_7494236 | 0.07 |
SOX15 |
SRY (sex determining region Y)-box 15 |
568 |
0.41 |
| chr12_118628413_118628612 | 0.07 |
TAOK3 |
TAO kinase 3 |
156 |
0.96 |
| chr13_64469784_64469935 | 0.07 |
ENSG00000202478 |
. |
111925 |
0.07 |
| chr2_179070529_179070680 | 0.07 |
OSBPL6 |
oxysterol binding protein-like 6 |
11228 |
0.22 |
| chr21_39678468_39679041 | 0.07 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
9906 |
0.29 |
| chr10_21803845_21803996 | 0.07 |
SKIDA1 |
SKI/DACH domain containing 1 |
2928 |
0.18 |
| chr9_23538750_23538901 | 0.07 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
196738 |
0.03 |
| chr2_51231032_51231183 | 0.07 |
NRXN1 |
neurexin 1 |
24304 |
0.21 |
| chr12_55371239_55371390 | 0.07 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
3600 |
0.28 |
| chr12_54746284_54746435 | 0.07 |
RP11-753H16.3 |
|
1086 |
0.24 |
| chr7_55509277_55509428 | 0.07 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
74418 |
0.1 |
| chr1_201605392_201605543 | 0.07 |
NAV1 |
neuron navigator 1 |
11910 |
0.15 |
| chr10_44427154_44427305 | 0.07 |
ENSG00000238957 |
. |
102759 |
0.08 |
| chr13_30276703_30277107 | 0.07 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
107080 |
0.07 |
| chr3_98616545_98616732 | 0.07 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
3377 |
0.22 |
| chr3_123518515_123518699 | 0.07 |
MYLK |
myosin light chain kinase |
5919 |
0.23 |
| chr1_245551646_245551892 | 0.07 |
ENSG00000221165 |
. |
18133 |
0.26 |
| chr2_207975517_207975668 | 0.07 |
ENSG00000253008 |
. |
795 |
0.64 |
| chr1_201426618_201426769 | 0.07 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
11619 |
0.15 |
| chr20_49638495_49638646 | 0.07 |
KCNG1 |
potassium voltage-gated channel, subfamily G, member 1 |
1047 |
0.51 |
| chr10_4284322_4284712 | 0.07 |
ENSG00000207124 |
. |
272627 |
0.02 |
| chr13_52922051_52922268 | 0.06 |
TPTE2P2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 pseudogene 2 |
50196 |
0.12 |
| chr2_11546902_11547171 | 0.06 |
AC099344.1 |
|
56891 |
0.1 |
| chr5_54127840_54127991 | 0.06 |
ENSG00000221073 |
. |
20861 |
0.24 |
| chr13_34184837_34185306 | 0.06 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
207115 |
0.02 |
| chr3_155109533_155109828 | 0.06 |
ENSG00000272096 |
. |
45932 |
0.16 |
| chr10_110881657_110881808 | 0.06 |
ENSG00000252574 |
. |
63033 |
0.15 |
| chr13_30095123_30095274 | 0.06 |
MTUS2-AS1 |
MTUS2 antisense RNA 1 |
30956 |
0.21 |
| chr1_156682649_156682800 | 0.06 |
RP11-66D17.5 |
|
242 |
0.84 |
| chr5_120534726_120534890 | 0.06 |
ENSG00000222609 |
. |
488273 |
0.01 |
| chr2_75784720_75784871 | 0.06 |
EVA1A |
eva-1 homolog A (C. elegans) |
3244 |
0.22 |
| chr14_102551048_102551199 | 0.06 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
1545 |
0.34 |
| chr2_231615765_231615916 | 0.06 |
ENSG00000201044 |
. |
1016 |
0.59 |
| chr1_146638747_146638898 | 0.06 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
5224 |
0.18 |
| chr4_2110491_2110642 | 0.06 |
RP11-317B7.3 |
|
30834 |
0.13 |
| chr12_889891_890042 | 0.06 |
ENSG00000221439 |
. |
333 |
0.9 |
| chr9_84302204_84302936 | 0.06 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
287 |
0.93 |
| chr2_47572405_47572655 | 0.06 |
EPCAM |
epithelial cell adhesion molecule |
233 |
0.93 |
| chr1_212874532_212874683 | 0.06 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
1280 |
0.42 |
| chr7_35756458_35756609 | 0.06 |
HERPUD2 |
HERPUD family member 2 |
21352 |
0.2 |
| chr10_119303944_119304573 | 0.06 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
321 |
0.88 |
| chr1_54422895_54423153 | 0.06 |
LRRC42 |
leucine rich repeat containing 42 |
10993 |
0.15 |
| chr9_14419303_14419454 | 0.06 |
NFIB |
nuclear factor I/B |
20396 |
0.26 |
| chr2_172388580_172388749 | 0.06 |
CYBRD1 |
cytochrome b reductase 1 |
9075 |
0.25 |
| chr6_137038589_137038792 | 0.06 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
61091 |
0.11 |
| chr5_50678440_50679177 | 0.06 |
ISL1 |
ISL LIM homeobox 1 |
113 |
0.79 |
| chr1_68663510_68663661 | 0.06 |
ENSG00000221203 |
. |
14292 |
0.21 |
| chr8_6400504_6400756 | 0.06 |
ANGPT2 |
angiopoietin 2 |
19935 |
0.24 |
| chr3_111673457_111673608 | 0.06 |
ABHD10 |
abhydrolase domain containing 10 |
24325 |
0.17 |
| chr2_19563220_19563383 | 0.06 |
OSR1 |
odd-skipped related transciption factor 1 |
4887 |
0.28 |
| chr12_10873934_10874085 | 0.06 |
YBX3 |
Y box binding protein 3 |
1897 |
0.32 |
| chr17_32583150_32583439 | 0.06 |
AC005549.3 |
Uncharacterized protein |
988 |
0.32 |
| chr7_121806775_121806926 | 0.06 |
AASS |
aminoadipate-semialdehyde synthase |
22516 |
0.24 |
| chr8_10872806_10873547 | 0.06 |
XKR6 |
XK, Kell blood group complex subunit-related family, member 6 |
13185 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.0 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |