Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
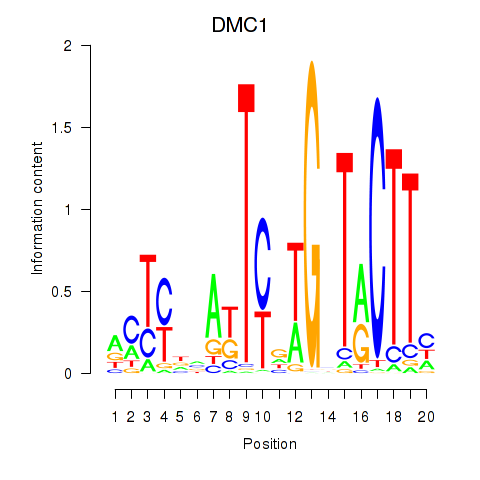
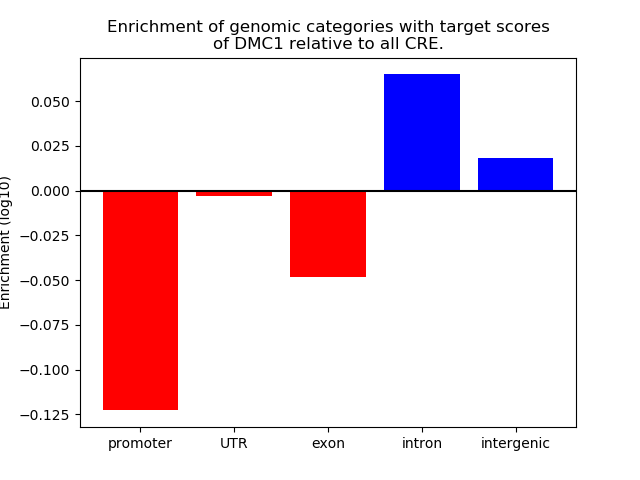
Results for DMC1
Z-value: 0.83
Transcription factors associated with DMC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMC1
|
ENSG00000100206.5 | DNA meiotic recombinase 1 |
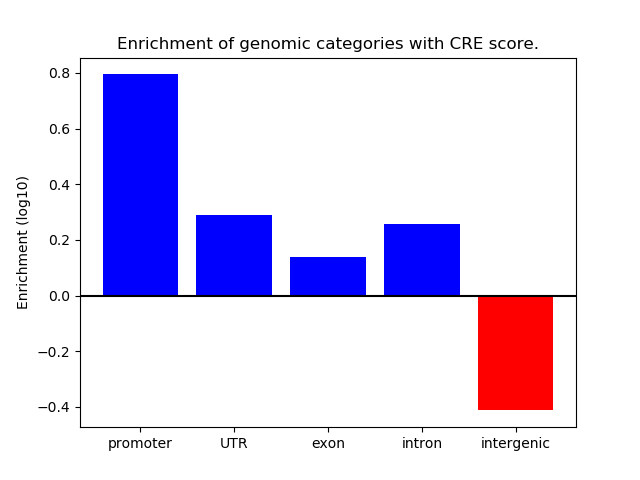
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr22_38966531_38967313 | DMC1 | 631 | 0.677684 | -0.59 | 9.3e-02 | Click! |
| chr22_38965920_38966142 | DMC1 | 105 | 0.960556 | -0.50 | 1.7e-01 | Click! |
| chr22_38965750_38965911 | DMC1 | 306 | 0.877472 | -0.46 | 2.1e-01 | Click! |
Activity of the DMC1 motif across conditions
Conditions sorted by the z-value of the DMC1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_36302041_36302306 | 0.36 |
RANBP3L |
RAN binding protein 3-like |
43 |
0.98 |
| chr4_183481639_183481857 | 0.35 |
ENSG00000252343 |
. |
11936 |
0.26 |
| chr7_130591247_130591471 | 0.33 |
ENSG00000226380 |
. |
29061 |
0.21 |
| chr1_63190926_63191187 | 0.32 |
RP11-230B22.1 |
|
36903 |
0.16 |
| chr15_80364416_80364822 | 0.31 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
287 |
0.93 |
| chr15_42082923_42083265 | 0.30 |
AC073657.1 |
Uncharacterized protein |
6865 |
0.12 |
| chr12_889349_889584 | 0.29 |
ENSG00000221439 |
. |
833 |
0.66 |
| chr2_102374666_102374817 | 0.29 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
38985 |
0.21 |
| chr17_74142136_74142287 | 0.29 |
RNF157-AS1 |
RNF157 antisense RNA 1 |
4614 |
0.13 |
| chr4_109347461_109347695 | 0.28 |
ENSG00000266046 |
. |
150718 |
0.04 |
| chr21_38352575_38353327 | 0.28 |
HLCS |
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
23 |
0.97 |
| chr5_38650925_38651503 | 0.28 |
LIFR |
leukemia inhibitory factor receptor alpha |
55708 |
0.13 |
| chr5_38844869_38845152 | 0.27 |
OSMR |
oncostatin M receptor |
950 |
0.68 |
| chr6_53480929_53481080 | 0.27 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
764 |
0.66 |
| chr2_227598429_227598675 | 0.27 |
IRS1 |
insulin receptor substrate 1 |
65923 |
0.11 |
| chr1_215739347_215739498 | 0.27 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
1313 |
0.59 |
| chr12_109248389_109248808 | 0.27 |
SSH1 |
slingshot protein phosphatase 1 |
2761 |
0.21 |
| chr7_97987211_97987389 | 0.26 |
RP11-307C18.1 |
|
35135 |
0.15 |
| chr8_59962698_59962849 | 0.26 |
RP11-328K2.1 |
|
57276 |
0.12 |
| chr4_90674169_90674343 | 0.26 |
RP11-115D19.1 |
|
71593 |
0.11 |
| chr3_152314332_152314547 | 0.25 |
RP11-362A9.3 |
|
99837 |
0.08 |
| chr18_7618536_7618784 | 0.25 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
50843 |
0.16 |
| chr19_50031610_50031873 | 0.24 |
RCN3 |
reticulocalbin 3, EF-hand calcium binding domain |
142 |
0.87 |
| chr4_187765290_187765672 | 0.24 |
ENSG00000252382 |
. |
13129 |
0.29 |
| chr1_83285095_83285395 | 0.24 |
LPHN2 |
latrophilin 2 |
839672 |
0.0 |
| chr20_32158375_32158526 | 0.24 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
8264 |
0.19 |
| chr7_22538999_22539909 | 0.24 |
STEAP1B |
STEAP family member 1B |
342 |
0.91 |
| chr3_135885490_135885641 | 0.24 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
27831 |
0.21 |
| chr1_243990574_243990792 | 0.23 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
15855 |
0.22 |
| chr14_99614015_99614166 | 0.23 |
AL162151.4 |
|
10663 |
0.28 |
| chr7_116151462_116151613 | 0.23 |
CAV2 |
caveolin 2 |
11703 |
0.15 |
| chr5_96000350_96000501 | 0.23 |
CAST |
calpastatin |
1668 |
0.37 |
| chr2_233497598_233498235 | 0.23 |
EFHD1 |
EF-hand domain family, member D1 |
22 |
0.97 |
| chr6_5498765_5499077 | 0.22 |
RP1-232P20.1 |
|
40613 |
0.2 |
| chr2_16817553_16817704 | 0.22 |
FAM49A |
family with sequence similarity 49, member A |
13284 |
0.3 |
| chr5_9534352_9534503 | 0.22 |
CTD-2201E9.2 |
|
11137 |
0.18 |
| chr12_116892640_116892791 | 0.22 |
ENSG00000264037 |
. |
26592 |
0.24 |
| chr3_190063110_190063261 | 0.21 |
CLDN1 |
claudin 1 |
22921 |
0.21 |
| chr10_63642613_63642764 | 0.21 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
18371 |
0.24 |
| chr10_97049571_97049740 | 0.21 |
PDLIM1 |
PDZ and LIM domain 1 |
1126 |
0.58 |
| chr22_44608120_44608309 | 0.21 |
PARVG |
parvin, gamma |
30473 |
0.22 |
| chr14_70140824_70141065 | 0.21 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
52673 |
0.13 |
| chr13_26623410_26623899 | 0.21 |
SHISA2 |
shisa family member 2 |
1515 |
0.52 |
| chr13_71953353_71953512 | 0.21 |
DACH1 |
dachshund homolog 1 (Drosophila) |
487475 |
0.01 |
| chr9_133721695_133722074 | 0.21 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
11431 |
0.21 |
| chr20_46110875_46111026 | 0.21 |
ENSG00000202186 |
. |
2175 |
0.28 |
| chr13_99880901_99881195 | 0.20 |
ENSG00000201793 |
. |
23086 |
0.16 |
| chr5_107767649_107767800 | 0.20 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
49925 |
0.19 |
| chr5_112823124_112823740 | 0.20 |
MCC |
mutated in colorectal cancers |
1095 |
0.49 |
| chr5_119951633_119951784 | 0.20 |
PRR16 |
proline rich 16 |
1075 |
0.68 |
| chrX_114468679_114469096 | 0.20 |
LRCH2 |
leucine-rich repeats and calponin homology (CH) domain containing 2 |
252 |
0.93 |
| chr18_70080364_70080515 | 0.20 |
CBLN2 |
cerebellin 2 precursor |
130350 |
0.06 |
| chr18_28741984_28742214 | 0.20 |
DSC1 |
desmocollin 1 |
720 |
0.67 |
| chr9_33510457_33510690 | 0.19 |
RP11-255A11.2 |
|
4198 |
0.19 |
| chr11_120202601_120202952 | 0.19 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
5011 |
0.19 |
| chr7_1443631_1443991 | 0.19 |
MICALL2 |
MICAL-like 2 |
55151 |
0.11 |
| chr7_107808332_107808483 | 0.19 |
NRCAM |
neuronal cell adhesion molecule |
7395 |
0.25 |
| chr22_37957123_37957770 | 0.19 |
CDC42EP1 |
CDC42 effector protein (Rho GTPase binding) 1 |
959 |
0.43 |
| chr7_15597655_15597806 | 0.19 |
AGMO |
alkylglycerol monooxygenase |
3910 |
0.36 |
| chr1_117342787_117343202 | 0.18 |
CD2 |
CD2 molecule |
45905 |
0.14 |
| chr15_48919611_48919986 | 0.18 |
FBN1 |
fibrillin 1 |
18120 |
0.26 |
| chr4_178167836_178167987 | 0.18 |
RP11-487E13.1 |
Uncharacterized protein |
2016 |
0.4 |
| chr12_57522507_57523183 | 0.18 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
38 |
0.64 |
| chr4_154125931_154126175 | 0.18 |
TRIM2 |
tripartite motif containing 2 |
435 |
0.87 |
| chr11_125036127_125036278 | 0.18 |
PKNOX2 |
PBX/knotted 1 homeobox 2 |
1552 |
0.29 |
| chr14_55741621_55741910 | 0.18 |
FBXO34 |
F-box protein 34 |
2893 |
0.32 |
| chr8_62504364_62504526 | 0.18 |
ENSG00000222898 |
. |
40126 |
0.18 |
| chr8_24799685_24800298 | 0.18 |
CTD-2168K21.2 |
|
14144 |
0.18 |
| chr3_74472777_74472928 | 0.18 |
CNTN3 |
contactin 3 (plasmacytoma associated) |
97439 |
0.09 |
| chr12_28125138_28125289 | 0.18 |
PTHLH |
parathyroid hormone-like hormone |
297 |
0.92 |
| chr14_55035520_55036036 | 0.18 |
SAMD4A |
sterile alpha motif domain containing 4A |
1141 |
0.56 |
| chr2_8422146_8422297 | 0.18 |
AC011747.7 |
|
393675 |
0.01 |
| chr8_69242814_69243988 | 0.18 |
C8orf34 |
chromosome 8 open reading frame 34 |
56 |
0.82 |
| chr10_44139678_44139829 | 0.18 |
ZNF32-AS1 |
ZNF32 antisense RNA 1 |
433 |
0.76 |
| chr4_77704559_77704807 | 0.18 |
RP11-359D14.3 |
|
18434 |
0.19 |
| chr2_88499667_88499997 | 0.17 |
THNSL2 |
threonine synthase-like 2 (S. cerevisiae) |
28853 |
0.14 |
| chr17_42293603_42293754 | 0.17 |
UBTF |
upstream binding transcription factor, RNA polymerase I |
2192 |
0.13 |
| chr1_42384263_42384523 | 0.17 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
15 |
0.99 |
| chr21_22573602_22573755 | 0.17 |
AP001136.2 |
|
25472 |
0.24 |
| chr9_8857014_8857547 | 0.17 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
496 |
0.84 |
| chr8_107738582_107738843 | 0.17 |
OXR1 |
oxidation resistance 1 |
299 |
0.9 |
| chr3_170148637_170148924 | 0.17 |
CLDN11 |
claudin 11 |
11925 |
0.25 |
| chr15_64635802_64635953 | 0.17 |
ENSG00000252774 |
. |
1139 |
0.39 |
| chr17_48714816_48714967 | 0.17 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
2673 |
0.17 |
| chr11_18341339_18341613 | 0.17 |
HPS5 |
Hermansky-Pudlak syndrome 5 |
2245 |
0.19 |
| chr3_15351793_15351990 | 0.17 |
ENSG00000238891 |
. |
6236 |
0.16 |
| chr17_1250998_1251685 | 0.17 |
YWHAE |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
13247 |
0.18 |
| chr9_130717953_130718104 | 0.17 |
FAM102A |
family with sequence similarity 102, member A |
5027 |
0.11 |
| chr8_41164878_41165029 | 0.17 |
SFRP1 |
secreted frizzled-related protein 1 |
444 |
0.81 |
| chr7_130680237_130680465 | 0.16 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
11483 |
0.25 |
| chr20_34743149_34743400 | 0.16 |
RP11-234K24.3 |
|
381 |
0.62 |
| chr6_155057921_155058172 | 0.16 |
SCAF8 |
SR-related CTD-associated factor 8 |
3496 |
0.32 |
| chr1_55701600_55701751 | 0.16 |
ENSG00000265822 |
. |
10361 |
0.22 |
| chr2_189434243_189434394 | 0.16 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
447 |
0.91 |
| chr14_68454519_68454814 | 0.16 |
CTD-2566J3.1 |
|
142247 |
0.04 |
| chr6_18043342_18043513 | 0.16 |
KIF13A |
kinesin family member 13A |
55573 |
0.13 |
| chr4_169616116_169616373 | 0.16 |
ENSG00000206613 |
. |
8377 |
0.21 |
| chr8_79965001_79965152 | 0.16 |
ENSG00000264969 |
. |
44522 |
0.21 |
| chr6_25193336_25193538 | 0.16 |
ENSG00000222373 |
. |
796 |
0.63 |
| chr15_94473358_94473509 | 0.16 |
ENSG00000222409 |
. |
33177 |
0.25 |
| chr5_40915264_40915466 | 0.15 |
ENSG00000253098 |
. |
4135 |
0.21 |
| chr3_30722457_30722608 | 0.15 |
RP11-1024P17.1 |
|
16621 |
0.27 |
| chr20_38091019_38091170 | 0.15 |
ENSG00000241840 |
. |
109692 |
0.07 |
| chr2_145594298_145594531 | 0.15 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
315793 |
0.01 |
| chr4_138480079_138480230 | 0.15 |
PCDH18 |
protocadherin 18 |
26506 |
0.28 |
| chr3_43682701_43682852 | 0.15 |
ANO10 |
anoctamin 10 |
19216 |
0.24 |
| chr4_138454899_138455069 | 0.15 |
PCDH18 |
protocadherin 18 |
1336 |
0.63 |
| chr1_226919798_226919959 | 0.15 |
ITPKB |
inositol-trisphosphate 3-kinase B |
5281 |
0.26 |
| chr2_85515912_85516063 | 0.15 |
ENSG00000221579 |
. |
10170 |
0.13 |
| chr22_26925865_26926016 | 0.15 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
10897 |
0.13 |
| chr4_54564237_54564388 | 0.15 |
RP11-317M11.1 |
|
2244 |
0.33 |
| chr12_111905255_111905406 | 0.15 |
ATXN2 |
ataxin 2 |
3034 |
0.24 |
| chr18_496799_496950 | 0.15 |
COLEC12 |
collectin sub-family member 12 |
3848 |
0.24 |
| chr2_44397964_44398115 | 0.15 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
1990 |
0.26 |
| chr6_80656707_80657785 | 0.15 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
51 |
0.98 |
| chr7_114649841_114649992 | 0.15 |
MDFIC |
MyoD family inhibitor domain containing |
75992 |
0.12 |
| chr18_2846122_2846273 | 0.15 |
EMILIN2 |
elastin microfibril interfacer 2 |
831 |
0.57 |
| chr17_762924_763100 | 0.15 |
NXN |
nucleoredoxin |
4325 |
0.18 |
| chr4_114677811_114678169 | 0.15 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
4234 |
0.36 |
| chr21_32591500_32591929 | 0.15 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
57369 |
0.16 |
| chr7_17341776_17341927 | 0.15 |
AC003075.4 |
|
2870 |
0.29 |
| chr19_49521308_49522757 | 0.15 |
LHB |
luteinizing hormone beta polypeptide |
1694 |
0.14 |
| chr8_95448497_95449318 | 0.15 |
FSBP |
fibrinogen silencer binding protein |
263 |
0.93 |
| chr2_87811764_87811915 | 0.15 |
RP11-1399P15.1 |
|
34286 |
0.22 |
| chrX_24497462_24497656 | 0.15 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
13986 |
0.21 |
| chr13_60348574_60349034 | 0.14 |
DIAPH3-AS1 |
DIAPH3 antisense RNA 1 |
238081 |
0.02 |
| chr1_6521246_6521397 | 0.14 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
4369 |
0.12 |
| chr6_46459858_46460009 | 0.14 |
RP11-795J1.1 |
|
144 |
0.62 |
| chr3_66502947_66503098 | 0.14 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
48334 |
0.18 |
| chr7_130663390_130663541 | 0.14 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
5403 |
0.28 |
| chrX_12029343_12029494 | 0.14 |
ENSG00000206792 |
. |
43627 |
0.19 |
| chr2_201375425_201375576 | 0.14 |
KCTD18 |
potassium channel tetramerization domain containing 18 |
709 |
0.45 |
| chr6_72103286_72103437 | 0.14 |
ENSG00000207827 |
. |
9963 |
0.21 |
| chr16_23144177_23144367 | 0.14 |
USP31 |
ubiquitin specific peptidase 31 |
16319 |
0.22 |
| chr5_52206164_52206315 | 0.14 |
ITGA2 |
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
78917 |
0.09 |
| chr13_63982821_63982972 | 0.14 |
LINC00395 |
long intergenic non-protein coding RNA 395 |
308255 |
0.01 |
| chr3_175244326_175245006 | 0.14 |
ENSG00000201648 |
. |
82601 |
0.1 |
| chr4_148602160_148602311 | 0.14 |
PRMT10 |
protein arginine methyltransferase 10 (putative) |
3041 |
0.27 |
| chrX_20157621_20157927 | 0.14 |
EIF1AX-AS1 |
EIF1AX antisense RNA 1 |
312 |
0.82 |
| chr7_104660604_104660779 | 0.14 |
KMT2E |
lysine (K)-specific methyltransferase 2E |
5783 |
0.17 |
| chr15_92499471_92499678 | 0.14 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
102223 |
0.08 |
| chr11_43665243_43665394 | 0.14 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
36941 |
0.17 |
| chr9_82427303_82427454 | 0.14 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
105697 |
0.08 |
| chr5_172306134_172306575 | 0.13 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
25866 |
0.16 |
| chr14_61224053_61224421 | 0.13 |
MNAT1 |
MNAT CDK-activating kinase assembly factor 1 |
22743 |
0.2 |
| chr17_61353070_61353290 | 0.13 |
RP11-269G24.2 |
|
37702 |
0.15 |
| chr5_77843393_77843700 | 0.13 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
1428 |
0.58 |
| chr11_118504420_118504703 | 0.13 |
AP002954.3 |
|
2777 |
0.17 |
| chr7_111201271_111201551 | 0.13 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
569 |
0.87 |
| chr9_3794388_3794539 | 0.13 |
RP11-252M18.3 |
|
81121 |
0.11 |
| chr2_144006322_144006473 | 0.13 |
RP11-190J23.1 |
|
76656 |
0.11 |
| chr11_14995465_14995616 | 0.13 |
CALCA |
calcitonin-related polypeptide alpha |
1640 |
0.48 |
| chr4_143481342_143481493 | 0.13 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
405 |
0.92 |
| chr8_131257957_131258144 | 0.13 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
1238 |
0.6 |
| chr8_131577804_131578063 | 0.13 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
122027 |
0.06 |
| chr9_95610203_95610354 | 0.13 |
ZNF484 |
zinc finger protein 484 |
8322 |
0.17 |
| chr1_23523630_23523879 | 0.13 |
HTR1D |
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
2532 |
0.23 |
| chr1_8686332_8686483 | 0.13 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
1968 |
0.44 |
| chr5_15533409_15533775 | 0.13 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
32045 |
0.23 |
| chr4_20289069_20289220 | 0.13 |
SLIT2 |
slit homolog 2 (Drosophila) |
32601 |
0.24 |
| chr3_116164058_116164214 | 0.13 |
LSAMP |
limbic system-associated membrane protein |
242 |
0.96 |
| chr2_181995547_181995698 | 0.13 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
148872 |
0.04 |
| chr6_132721617_132721768 | 0.13 |
MOXD1 |
monooxygenase, DBH-like 1 |
922 |
0.69 |
| chr8_99952075_99952653 | 0.13 |
STK3 |
serine/threonine kinase 3 |
2435 |
0.25 |
| chr3_168744118_168744269 | 0.13 |
MECOM |
MDS1 and EVI1 complex locus |
101629 |
0.08 |
| chr10_35079377_35079528 | 0.13 |
PARD3 |
par-3 family cell polarity regulator |
24797 |
0.2 |
| chr4_57156140_57156443 | 0.13 |
ENSG00000252489 |
. |
1541 |
0.39 |
| chr5_35855125_35855276 | 0.12 |
IL7R |
interleukin 7 receptor |
1197 |
0.5 |
| chr6_160509984_160510135 | 0.12 |
IGF2R |
insulin-like growth factor 2 receptor |
3966 |
0.27 |
| chr19_4473895_4474732 | 0.12 |
HDGFRP2 |
Hepatoma-derived growth factor-related protein 2 |
576 |
0.53 |
| chr1_61598053_61598265 | 0.12 |
RP4-802A10.1 |
|
7754 |
0.25 |
| chr7_80543186_80543337 | 0.12 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
5238 |
0.35 |
| chr3_12925603_12925754 | 0.12 |
AC034198.7 |
|
33520 |
0.13 |
| chr5_16827159_16827310 | 0.12 |
ENSG00000253093 |
. |
26869 |
0.22 |
| chr12_70937490_70937641 | 0.12 |
RP11-588H23.3 |
|
23579 |
0.22 |
| chr1_103573283_103573505 | 0.12 |
COL11A1 |
collagen, type XI, alpha 1 |
340 |
0.94 |
| chr4_41122983_41123211 | 0.12 |
ENSG00000207198 |
. |
7138 |
0.23 |
| chr14_91525121_91525272 | 0.12 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
1282 |
0.35 |
| chr2_115419198_115419381 | 0.12 |
DPP10-AS3 |
DPP10 antisense RNA 3 |
171836 |
0.03 |
| chr4_90056009_90056160 | 0.12 |
ENSG00000212226 |
. |
4081 |
0.27 |
| chr22_36232996_36233711 | 0.12 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
2912 |
0.37 |
| chr2_145261378_145261529 | 0.12 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
13662 |
0.24 |
| chr5_112481657_112481808 | 0.12 |
MCC |
mutated in colorectal cancers |
88642 |
0.09 |
| chr6_108108889_108109040 | 0.12 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
15509 |
0.27 |
| chr1_193509624_193509775 | 0.12 |
ENSG00000252241 |
. |
191375 |
0.03 |
| chr21_36359858_36360101 | 0.12 |
RUNX1 |
runt-related transcription factor 1 |
61483 |
0.16 |
| chr2_112195991_112196142 | 0.12 |
ENSG00000266139 |
. |
117398 |
0.07 |
| chr7_137638804_137638955 | 0.12 |
AC022173.2 |
|
785 |
0.68 |
| chr13_41487964_41488115 | 0.12 |
SUGT1P3 |
SUGT1 pseudogene 3 |
7808 |
0.19 |
| chr20_22756193_22756438 | 0.12 |
ENSG00000265151 |
. |
42376 |
0.2 |
| chr9_123693983_123694176 | 0.12 |
TRAF1 |
TNF receptor-associated factor 1 |
2628 |
0.31 |
| chr3_8493679_8493830 | 0.12 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
49530 |
0.14 |
| chr7_25431142_25431293 | 0.12 |
ENSG00000202233 |
. |
126942 |
0.05 |
| chr6_73456305_73456456 | 0.12 |
KCNQ5-IT1 |
KCNQ5 intronic transcript 1 (non-protein coding) |
116157 |
0.06 |
| chr6_27440042_27440937 | 0.12 |
ZNF184 |
zinc finger protein 184 |
29 |
0.98 |
| chr12_78836151_78836302 | 0.12 |
NAV3 |
neuron navigator 3 |
243370 |
0.02 |
| chr11_86085513_86085741 | 0.12 |
CCDC81 |
coiled-coil domain containing 81 |
151 |
0.96 |
| chr3_87487041_87487197 | 0.12 |
POU1F1 |
POU class 1 homeobox 1 |
161382 |
0.04 |
| chr13_52217695_52217893 | 0.12 |
ENSG00000242893 |
. |
49443 |
0.11 |
| chr18_66506011_66506162 | 0.12 |
CCDC102B |
coiled-coil domain containing 102B |
2085 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.0 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.2 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0010875 | positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0002837 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0072191 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |