Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
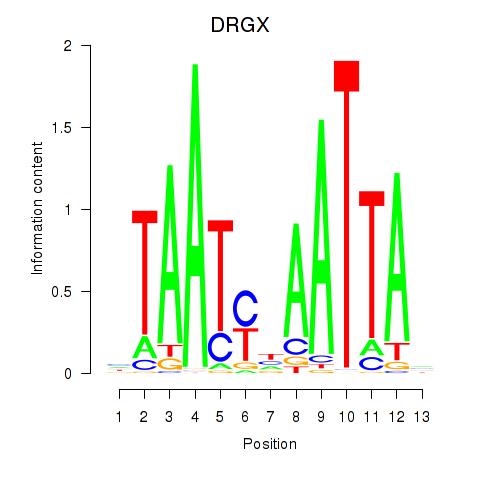
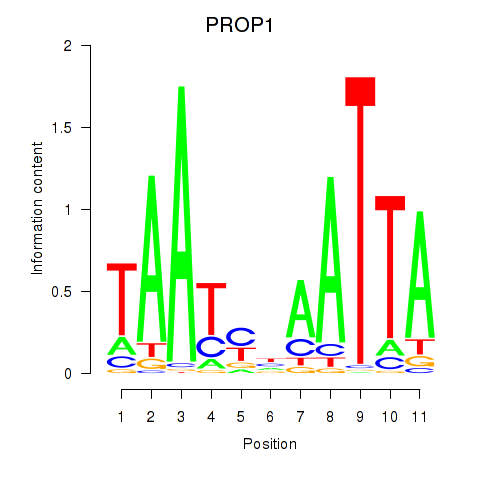
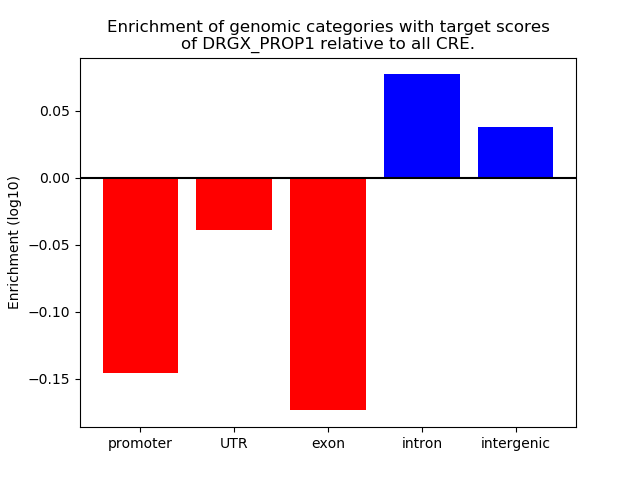
Results for DRGX_PROP1
Z-value: 0.76
Transcription factors associated with DRGX_PROP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DRGX
|
ENSG00000165606.4 | dorsal root ganglia homeobox |
|
PROP1
|
ENSG00000175325.2 | PROP paired-like homeobox 1 |
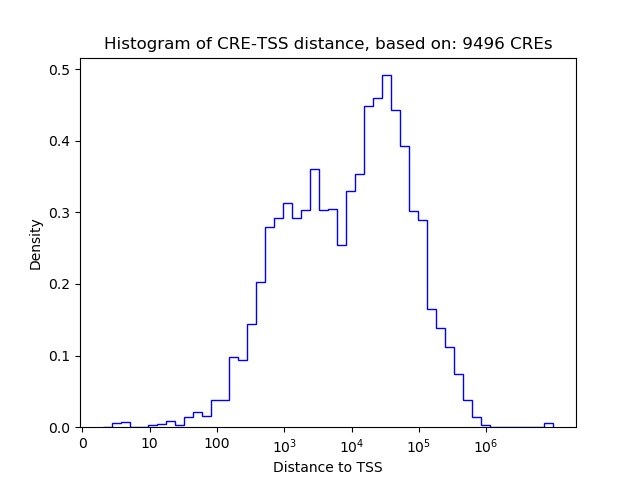
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_50606076_50606780 | DRGX | 2931 | 0.266552 | 0.86 | 2.8e-03 | Click! |
| chr5_177411424_177411575 | PROP1 | 11744 | 0.147987 | 0.68 | 4.3e-02 | Click! |
| chr5_177411774_177412187 | PROP1 | 11263 | 0.149068 | 0.60 | 8.7e-02 | Click! |
| chr5_177412450_177412601 | PROP1 | 10718 | 0.150331 | 0.20 | 6.0e-01 | Click! |
Activity of the DRGX_PROP1 motif across conditions
Conditions sorted by the z-value of the DRGX_PROP1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
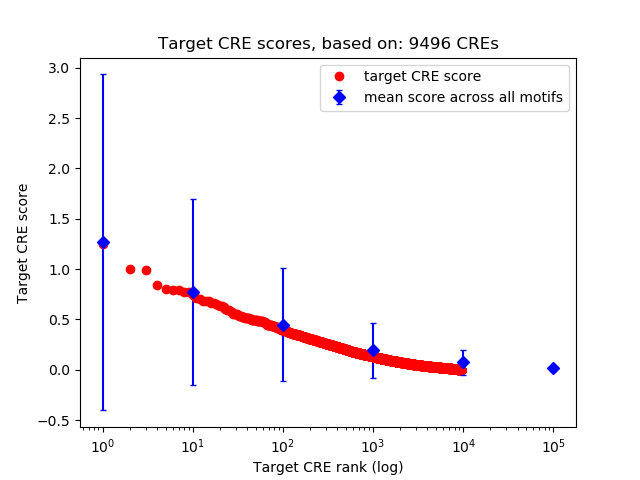
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_54968513_54968925 | 1.25 |
IRX5 |
iroquois homeobox 5 |
2735 |
0.43 |
| chr22_36310772_36312004 | 1.00 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
23557 |
0.25 |
| chr4_187647302_187647732 | 0.99 |
FAT1 |
FAT atypical cadherin 1 |
359 |
0.93 |
| chr15_37179413_37179956 | 0.85 |
ENSG00000212511 |
. |
34843 |
0.22 |
| chr11_131780111_131780399 | 0.81 |
NTM |
neurotrimin |
642 |
0.77 |
| chr1_120217864_120218312 | 0.80 |
ZNF697 |
zinc finger protein 697 |
27692 |
0.16 |
| chr1_240250348_240250507 | 0.79 |
FMN2 |
formin 2 |
4753 |
0.3 |
| chr12_49762046_49762218 | 0.77 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
802 |
0.53 |
| chr1_179111271_179111703 | 0.77 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
692 |
0.72 |
| chr15_38545953_38546360 | 0.75 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
774 |
0.79 |
| chr7_27202641_27202952 | 0.71 |
HOXA9 |
homeobox A9 |
2349 |
0.09 |
| chr11_121593168_121593875 | 0.70 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
132393 |
0.05 |
| chr4_159091107_159091452 | 0.69 |
FAM198B |
family with sequence similarity 198, member B |
251 |
0.77 |
| chr2_54682726_54683239 | 0.68 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
440 |
0.88 |
| chr2_197852125_197852688 | 0.68 |
ANKRD44 |
ankyrin repeat domain 44 |
12820 |
0.25 |
| chr16_53244996_53245358 | 0.67 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
2814 |
0.29 |
| chr12_59311795_59312096 | 0.67 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1382 |
0.46 |
| chr3_79066923_79067090 | 0.65 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
688 |
0.81 |
| chr21_28214648_28215240 | 0.64 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
393 |
0.9 |
| chr22_46462951_46463814 | 0.63 |
RP6-109B7.4 |
|
2389 |
0.16 |
| chr4_138045655_138046009 | 0.63 |
ENSG00000222526 |
. |
103450 |
0.09 |
| chr14_45433796_45433989 | 0.62 |
FAM179B |
family with sequence similarity 179, member B |
2450 |
0.23 |
| chr13_38172411_38172686 | 0.60 |
POSTN |
periostin, osteoblast specific factor |
315 |
0.95 |
| chr18_60385543_60385694 | 0.59 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
2935 |
0.28 |
| chr9_12777034_12777185 | 0.59 |
LURAP1L |
leucine rich adaptor protein 1-like |
2089 |
0.33 |
| chr12_3308931_3310293 | 0.58 |
TSPAN9 |
tetraspanin 9 |
729 |
0.75 |
| chr3_88191159_88191414 | 0.58 |
ZNF654 |
zinc finger protein 654 |
3032 |
0.24 |
| chr17_79374852_79375003 | 0.56 |
ENSG00000266392 |
. |
349 |
0.75 |
| chr4_54187854_54188041 | 0.55 |
SCFD2 |
sec1 family domain containing 2 |
44281 |
0.15 |
| chr2_16831792_16832248 | 0.55 |
FAM49A |
family with sequence similarity 49, member A |
15076 |
0.3 |
| chr2_151334124_151334616 | 0.55 |
RND3 |
Rho family GTPase 3 |
7526 |
0.34 |
| chr3_79066759_79066910 | 0.54 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
860 |
0.75 |
| chr6_56405555_56405706 | 0.54 |
DST |
dystonin |
66842 |
0.13 |
| chr20_56706361_56706780 | 0.53 |
C20orf85 |
chromosome 20 open reading frame 85 |
19390 |
0.21 |
| chr8_24867956_24868211 | 0.53 |
CTD-2168K21.2 |
|
53948 |
0.14 |
| chr8_49468858_49469448 | 0.52 |
RP11-770E5.1 |
|
5026 |
0.35 |
| chr16_24999320_24999715 | 0.52 |
ARHGAP17 |
Rho GTPase activating protein 17 |
27135 |
0.22 |
| chr5_94416178_94416653 | 0.52 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
771 |
0.78 |
| chr6_169651321_169651472 | 0.51 |
THBS2 |
thrombospondin 2 |
850 |
0.72 |
| chr14_71067961_71068112 | 0.51 |
MED6 |
mediator complex subunit 6 |
652 |
0.7 |
| chr6_116833589_116833880 | 0.51 |
TRAPPC3L |
trafficking protein particle complex 3-like |
228 |
0.83 |
| chr15_62917508_62917659 | 0.51 |
RP11-625H11.1 |
Uncharacterized protein |
19797 |
0.2 |
| chr17_35851874_35852313 | 0.50 |
DUSP14 |
dual specificity phosphatase 14 |
523 |
0.79 |
| chrX_106241917_106242068 | 0.50 |
MORC4 |
MORC family CW-type zinc finger 4 |
1316 |
0.53 |
| chr1_210424215_210424840 | 0.50 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
17135 |
0.21 |
| chr13_52979416_52980324 | 0.50 |
THSD1 |
thrombospondin, type I, domain containing 1 |
393 |
0.85 |
| chr3_12328729_12329240 | 0.49 |
PPARG |
peroxisome proliferator-activated receptor gamma |
97 |
0.98 |
| chr12_26379464_26379833 | 0.49 |
SSPN |
sarcospan |
31042 |
0.17 |
| chr15_96897591_96898313 | 0.49 |
AC087477.1 |
Uncharacterized protein |
6535 |
0.18 |
| chr4_187646786_187647042 | 0.49 |
FAT1 |
FAT atypical cadherin 1 |
962 |
0.7 |
| chr12_64581322_64581595 | 0.49 |
ENSG00000212298 |
. |
17115 |
0.15 |
| chr1_157013915_157014350 | 0.49 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
733 |
0.68 |
| chr1_89197420_89197765 | 0.49 |
PKN2 |
protein kinase N2 |
39546 |
0.16 |
| chr6_27859319_27859470 | 0.49 |
HIST1H3J |
histone cluster 1, H3j |
824 |
0.31 |
| chr1_179110399_179110598 | 0.49 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
1681 |
0.39 |
| chr4_89514978_89515129 | 0.49 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
524 |
0.77 |
| chr2_101437792_101437972 | 0.48 |
NPAS2 |
neuronal PAS domain protein 2 |
395 |
0.8 |
| chr1_95436764_95437146 | 0.48 |
RP4-639F20.1 |
|
10074 |
0.2 |
| chr8_18577899_18578296 | 0.48 |
RP11-161I2.1 |
|
318 |
0.92 |
| chr15_36043742_36043931 | 0.48 |
ENSG00000265098 |
. |
175221 |
0.03 |
| chr11_133990232_133990720 | 0.48 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
47891 |
0.12 |
| chr4_48332309_48332534 | 0.47 |
SLAIN2 |
SLAIN motif family, member 2 |
10918 |
0.24 |
| chr7_13985407_13985644 | 0.47 |
ETV1 |
ets variant 1 |
40541 |
0.2 |
| chr7_25901481_25901888 | 0.47 |
ENSG00000199085 |
. |
87922 |
0.1 |
| chr12_28121780_28121931 | 0.47 |
PTHLH |
parathyroid hormone-like hormone |
572 |
0.79 |
| chr5_54456456_54456818 | 0.45 |
GPX8 |
glutathione peroxidase 8 (putative) |
639 |
0.58 |
| chr18_7568558_7569022 | 0.44 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
973 |
0.7 |
| chr20_34463775_34463999 | 0.44 |
ENSG00000201221 |
. |
11694 |
0.14 |
| chr6_125420988_125421372 | 0.44 |
TPD52L1 |
tumor protein D52-like 1 |
19015 |
0.24 |
| chr14_51909020_51909357 | 0.44 |
FRMD6 |
FERM domain containing 6 |
46667 |
0.16 |
| chr1_212643718_212643913 | 0.44 |
NENF |
neudesin neurotrophic factor |
37586 |
0.14 |
| chr10_33620140_33620362 | 0.44 |
NRP1 |
neuropilin 1 |
3059 |
0.34 |
| chr12_75841002_75841191 | 0.44 |
GLIPR1 |
GLI pathogenesis-related 1 |
33364 |
0.14 |
| chr4_48701412_48701950 | 0.44 |
FRYL |
FRY-like |
18493 |
0.24 |
| chr18_52988378_52989650 | 0.44 |
TCF4 |
transcription factor 4 |
203 |
0.97 |
| chr19_30719241_30719606 | 0.44 |
ZNF536 |
zinc finger protein 536 |
2 |
0.73 |
| chr1_54199074_54199253 | 0.43 |
GLIS1 |
GLIS family zinc finger 1 |
714 |
0.69 |
| chr6_16737749_16737900 | 0.43 |
RP1-151F17.1 |
|
23545 |
0.22 |
| chr13_40139928_40140149 | 0.43 |
LHFP |
lipoma HMGIC fusion partner |
37270 |
0.18 |
| chr17_33391542_33391948 | 0.43 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
874 |
0.45 |
| chr5_53753293_53753597 | 0.43 |
HSPB3 |
heat shock 27kDa protein 3 |
2000 |
0.44 |
| chr9_5631788_5632177 | 0.43 |
KIAA1432 |
KIAA1432 |
2655 |
0.36 |
| chr11_35188701_35188984 | 0.42 |
CD44 |
CD44 molecule (Indian blood group) |
9276 |
0.17 |
| chr3_136505464_136505764 | 0.42 |
SLC35G2 |
solute carrier family 35, member G2 |
32247 |
0.12 |
| chr12_75784171_75784441 | 0.42 |
CAPS2 |
calcyphosine 2 |
375 |
0.6 |
| chrX_114829080_114829277 | 0.42 |
PLS3 |
plastin 3 |
1313 |
0.48 |
| chr21_38219417_38219579 | 0.42 |
ENSG00000199806 |
. |
4713 |
0.25 |
| chr8_89338531_89338740 | 0.42 |
RP11-586K2.1 |
|
430 |
0.76 |
| chr1_184858090_184858241 | 0.42 |
ENSG00000252222 |
. |
67542 |
0.11 |
| chr7_116141197_116141631 | 0.42 |
CAV2 |
caveolin 2 |
1580 |
0.29 |
| chr15_98506044_98506195 | 0.41 |
ARRDC4 |
arrestin domain containing 4 |
2191 |
0.38 |
| chr12_47219854_47220069 | 0.41 |
SLC38A4 |
solute carrier family 38, member 4 |
181 |
0.97 |
| chr5_142623144_142623555 | 0.41 |
ARHGAP26 |
Rho GTPase activating protein 26 |
36584 |
0.19 |
| chr1_95330057_95330663 | 0.41 |
SLC44A3 |
solute carrier family 44, member 3 |
2527 |
0.28 |
| chr6_121758427_121758731 | 0.40 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
1741 |
0.35 |
| chr4_39938923_39939074 | 0.40 |
ENSG00000252970 |
. |
625 |
0.75 |
| chr21_36264537_36264739 | 0.40 |
RUNX1 |
runt-related transcription factor 1 |
2551 |
0.42 |
| chr7_32099128_32099768 | 0.40 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
11013 |
0.32 |
| chr9_708078_708584 | 0.40 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
1442 |
0.47 |
| chr8_16576311_16576462 | 0.40 |
ENSG00000264092 |
. |
144619 |
0.05 |
| chr8_37678948_37679463 | 0.39 |
GPR124 |
G protein-coupled receptor 124 |
24431 |
0.12 |
| chr16_86599945_86600151 | 0.39 |
FOXC2 |
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
809 |
0.48 |
| chr19_13136767_13137028 | 0.39 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
1086 |
0.34 |
| chr10_12095697_12095857 | 0.39 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
10754 |
0.17 |
| chr17_67137246_67137397 | 0.39 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
694 |
0.69 |
| chr1_68697735_68698156 | 0.39 |
WLS |
wntless Wnt ligand secretion mediator |
18 |
0.98 |
| chr18_74202036_74202284 | 0.39 |
ZNF516 |
zinc finger protein 516 |
575 |
0.66 |
| chr4_87935725_87935950 | 0.39 |
AFF1 |
AF4/FMR2 family, member 1 |
7416 |
0.29 |
| chr18_30563491_30563642 | 0.38 |
RP11-680N20.1 |
|
29527 |
0.24 |
| chr20_10502111_10502382 | 0.38 |
SLX4IP |
SLX4 interacting protein |
86295 |
0.09 |
| chr7_82188634_82189045 | 0.38 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
115725 |
0.07 |
| chr2_36924982_36925262 | 0.38 |
VIT |
vitrin |
1102 |
0.6 |
| chr1_119529068_119529220 | 0.38 |
TBX15 |
T-box 15 |
1284 |
0.57 |
| chr5_88120627_88121096 | 0.37 |
MEF2C |
myocyte enhancer factor 2C |
704 |
0.78 |
| chr2_64926239_64926446 | 0.37 |
SERTAD2 |
SERTA domain containing 2 |
45295 |
0.15 |
| chr8_16689434_16690090 | 0.37 |
ENSG00000264092 |
. |
31243 |
0.24 |
| chr3_181434209_181434555 | 0.37 |
SOX2 |
SRY (sex determining region Y)-box 2 |
4660 |
0.26 |
| chr2_200446222_200446758 | 0.37 |
SATB2 |
SATB homeobox 2 |
110501 |
0.07 |
| chr4_15010597_15010748 | 0.37 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
5112 |
0.32 |
| chr10_119132010_119132432 | 0.37 |
PDZD8 |
PDZ domain containing 8 |
2757 |
0.3 |
| chr5_88027797_88028081 | 0.37 |
CTC-467M3.2 |
|
39471 |
0.16 |
| chr11_102959875_102960225 | 0.37 |
DCUN1D5 |
DCN1, defective in cullin neddylation 1, domain containing 5 |
3 |
0.98 |
| chr9_80409118_80409269 | 0.37 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
28722 |
0.25 |
| chr18_67713592_67713743 | 0.37 |
RTTN |
rotatin |
25713 |
0.25 |
| chr15_56206090_56206310 | 0.36 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
1652 |
0.45 |
| chr3_136677766_136677917 | 0.36 |
IL20RB |
interleukin 20 receptor beta |
990 |
0.56 |
| chr1_39860957_39861272 | 0.36 |
KIAA0754 |
KIAA0754 |
15037 |
0.18 |
| chr8_70406794_70407007 | 0.36 |
SULF1 |
sulfatase 1 |
1872 |
0.5 |
| chr7_123390243_123390394 | 0.36 |
RP11-390E23.6 |
|
1196 |
0.34 |
| chr1_23696524_23696675 | 0.36 |
ENSG00000201405 |
. |
148 |
0.82 |
| chr12_66220556_66220804 | 0.36 |
HMGA2 |
high mobility group AT-hook 2 |
1777 |
0.39 |
| chr13_39945288_39945439 | 0.36 |
ENSG00000238408 |
. |
75990 |
0.12 |
| chr2_189247602_189247926 | 0.36 |
ENSG00000207951 |
. |
85545 |
0.09 |
| chr12_78336292_78336638 | 0.35 |
NAV3 |
neuron navigator 3 |
23591 |
0.28 |
| chr22_35778076_35778227 | 0.35 |
HMOX1 |
heme oxygenase (decycling) 1 |
1323 |
0.43 |
| chr10_119303944_119304573 | 0.35 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
321 |
0.88 |
| chr4_61665381_61665532 | 0.35 |
ENSG00000265829 |
. |
122881 |
0.06 |
| chr6_26577102_26577344 | 0.35 |
ABT1 |
activator of basal transcription 1 |
19957 |
0.15 |
| chr5_53386895_53387046 | 0.35 |
ENSG00000265421 |
. |
15557 |
0.24 |
| chr9_96902091_96902301 | 0.35 |
ENSG00000199165 |
. |
36038 |
0.13 |
| chr2_237479145_237479357 | 0.35 |
ACKR3 |
atypical chemokine receptor 3 |
967 |
0.68 |
| chr1_86048991_86049142 | 0.35 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
2622 |
0.26 |
| chr2_171381865_171382027 | 0.35 |
AC007277.3 |
|
118508 |
0.06 |
| chr5_139418219_139418461 | 0.35 |
NRG2 |
neuregulin 2 |
4314 |
0.21 |
| chr2_12842465_12842741 | 0.35 |
TRIB2 |
tribbles pseudokinase 2 |
14412 |
0.26 |
| chr3_126500993_126501380 | 0.35 |
CHCHD6 |
coiled-coil-helix-coiled-coil-helix domain containing 6 |
51779 |
0.16 |
| chr22_44722319_44722530 | 0.35 |
KIAA1644 |
KIAA1644 |
13693 |
0.24 |
| chr4_114388823_114389059 | 0.35 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
46057 |
0.14 |
| chr15_38548551_38548873 | 0.35 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
3330 |
0.39 |
| chr10_54202834_54203158 | 0.34 |
RP11-556E13.1 |
|
117869 |
0.06 |
| chr5_61389615_61389794 | 0.34 |
ENSG00000251983 |
. |
52788 |
0.18 |
| chr21_32715523_32716836 | 0.34 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
415 |
0.91 |
| chr6_111582018_111582254 | 0.34 |
KIAA1919 |
KIAA1919 |
1585 |
0.29 |
| chr13_21388856_21389007 | 0.34 |
N6AMT2 |
N-6 adenine-specific DNA methyltransferase 2 (putative) |
40843 |
0.16 |
| chr2_98765349_98765752 | 0.34 |
VWA3B |
von Willebrand factor A domain containing 3B |
61851 |
0.13 |
| chr14_32037334_32037518 | 0.34 |
NUBPL |
nucleotide binding protein-like |
6596 |
0.17 |
| chr4_184040476_184040627 | 0.34 |
WWC2 |
WW and C2 domain containing 2 |
19906 |
0.19 |
| chr4_114705923_114706074 | 0.34 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
22915 |
0.27 |
| chr8_107738861_107739281 | 0.34 |
OXR1 |
oxidation resistance 1 |
658 |
0.71 |
| chr9_123919065_123919216 | 0.34 |
CNTRL |
centriolin |
981 |
0.55 |
| chr12_125170011_125170375 | 0.33 |
NCOR2 |
nuclear receptor corepressor 2 |
118183 |
0.06 |
| chr7_78911351_78911863 | 0.33 |
ENSG00000212482 |
. |
61081 |
0.15 |
| chr7_116222889_116223134 | 0.33 |
AC006159.4 |
|
11723 |
0.18 |
| chr4_23890359_23890765 | 0.33 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
1096 |
0.66 |
| chr14_50692436_50692613 | 0.33 |
SOS2 |
son of sevenless homolog 2 (Drosophila) |
5503 |
0.22 |
| chr5_66254399_66254664 | 0.33 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
167 |
0.97 |
| chr14_35179773_35180008 | 0.33 |
CFL2 |
cofilin 2 (muscle) |
3144 |
0.26 |
| chr11_94503028_94503179 | 0.33 |
AMOTL1 |
angiomotin like 1 |
1566 |
0.45 |
| chr4_102277312_102277561 | 0.33 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
8001 |
0.2 |
| chr14_29254002_29254283 | 0.33 |
RP11-966I7.2 |
|
851 |
0.56 |
| chr2_197831476_197831662 | 0.33 |
ANKRD44 |
ankyrin repeat domain 44 |
33657 |
0.18 |
| chr17_27719677_27719955 | 0.33 |
TAOK1 |
TAO kinase 1 |
1867 |
0.18 |
| chr12_78334931_78335200 | 0.33 |
NAV3 |
neuron navigator 3 |
24991 |
0.28 |
| chr14_79747812_79748200 | 0.33 |
NRXN3 |
neurexin 3 |
1757 |
0.44 |
| chr9_97666762_97667035 | 0.32 |
RP11-49O14.2 |
|
4187 |
0.23 |
| chr1_68297324_68297475 | 0.32 |
GNG12-AS1 |
GNG12 antisense RNA 1 |
587 |
0.71 |
| chr21_17679599_17679750 | 0.32 |
ENSG00000201025 |
. |
22585 |
0.28 |
| chr8_32441619_32441833 | 0.32 |
NRG1 |
neuregulin 1 |
21440 |
0.28 |
| chr1_71831333_71831497 | 0.32 |
ENSG00000229956 |
. |
35990 |
0.23 |
| chr4_81192022_81192173 | 0.32 |
FGF5 |
fibroblast growth factor 5 |
4304 |
0.29 |
| chr2_110656312_110656910 | 0.32 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
343 |
0.93 |
| chr10_29916878_29917029 | 0.32 |
SVIL |
supervillin |
6948 |
0.21 |
| chr12_8814882_8815352 | 0.32 |
MFAP5 |
microfibrillar associated protein 5 |
98 |
0.95 |
| chr6_10409161_10409312 | 0.32 |
TFAP2A-AS1 |
TFAP2A antisense RNA 1 |
337 |
0.8 |
| chr10_33665800_33666194 | 0.32 |
NRP1 |
neuropilin 1 |
40807 |
0.2 |
| chr15_56205778_56205950 | 0.32 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
1988 |
0.4 |
| chr7_7197772_7198055 | 0.32 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
1348 |
0.45 |
| chr4_176692255_176692416 | 0.32 |
GPM6A |
glycoprotein M6A |
16203 |
0.23 |
| chr15_25638050_25638201 | 0.32 |
UBE3A |
ubiquitin protein ligase E3A |
12528 |
0.11 |
| chr14_79426395_79426572 | 0.32 |
CTD-2243E23.1 |
|
114622 |
0.07 |
| chr4_54424539_54424751 | 0.31 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
261 |
0.93 |
| chr20_14316163_14316462 | 0.31 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
1942 |
0.4 |
| chr3_151694269_151694458 | 0.31 |
RP11-454C18.2 |
|
48400 |
0.17 |
| chr6_3180552_3180703 | 0.31 |
TUBB2A |
tubulin, beta 2A class IIa |
22867 |
0.13 |
| chr20_7148010_7148161 | 0.31 |
ENSG00000251833 |
. |
306424 |
0.01 |
| chr18_56532811_56532962 | 0.31 |
ZNF532 |
zinc finger protein 532 |
100 |
0.97 |
| chr2_12928938_12929131 | 0.31 |
ENSG00000264370 |
. |
51541 |
0.17 |
| chr2_109271526_109272143 | 0.31 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
325 |
0.91 |
| chr6_125684129_125684280 | 0.31 |
RP11-735G4.1 |
|
11266 |
0.27 |
| chr4_170069363_170069514 | 0.31 |
RP11-327O17.2 |
|
53507 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.6 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.2 | 0.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.4 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.3 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.3 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.1 | 0.6 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.3 | GO:0010665 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.2 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.6 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.3 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.2 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0032823 | natural killer cell differentiation(GO:0001779) regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0051940 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.2 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.4 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.8 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.0 | 0.1 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0060900 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.2 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.4 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0090030 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0001974 | blood vessel remodeling(GO:0001974) |
| 0.0 | 1.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.1 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.0 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:0021930 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.0 | GO:0003211 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.0 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0048333 | mesodermal cell differentiation(GO:0048333) |
| 0.0 | 0.4 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.0 | GO:0072110 | cellular response to heparin(GO:0071504) glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.0 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.2 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.4 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 2.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 0.5 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.2 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.4 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.4 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |