Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
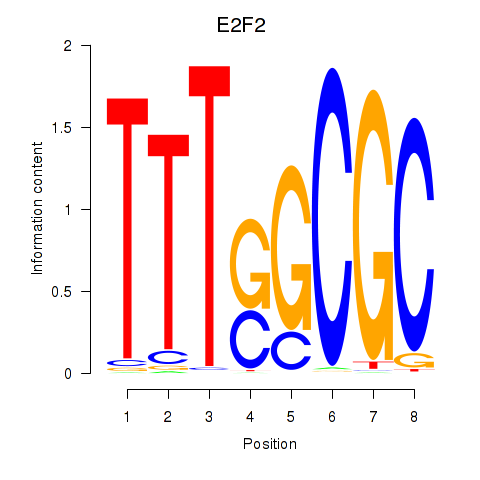
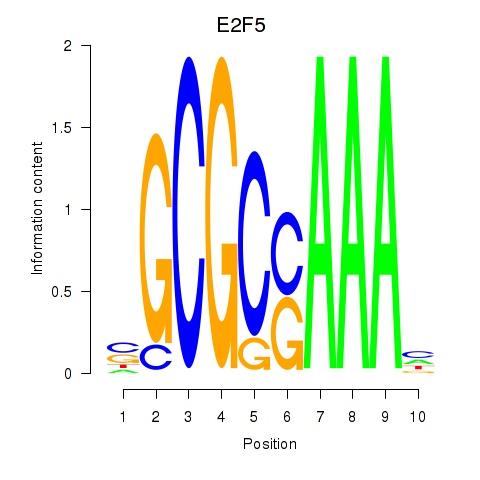
Results for E2F2_E2F5
Z-value: 0.82
Transcription factors associated with E2F2_E2F5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F2
|
ENSG00000007968.6 | E2F transcription factor 2 |
|
E2F5
|
ENSG00000133740.6 | E2F transcription factor 5 |
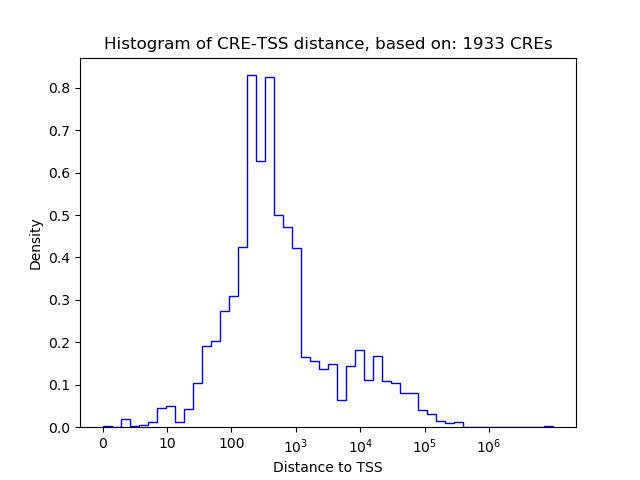
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_23854853_23855052 | E2F2 | 2760 | 0.249146 | 0.70 | 3.6e-02 | Click! |
| chr1_23857954_23858302 | E2F2 | 416 | 0.833272 | 0.70 | 3.6e-02 | Click! |
| chr1_23866506_23866674 | E2F2 | 8878 | 0.175383 | 0.69 | 3.8e-02 | Click! |
| chr1_23856502_23856679 | E2F2 | 1122 | 0.484736 | 0.62 | 7.6e-02 | Click! |
| chr1_23866282_23866470 | E2F2 | 8664 | 0.176025 | 0.61 | 8.1e-02 | Click! |
| chr8_86112985_86113136 | E2F5 | 5338 | 0.140923 | -0.44 | 2.4e-01 | Click! |
| chr8_86089338_86089536 | E2F5 | 23 | 0.624274 | 0.41 | 2.7e-01 | Click! |
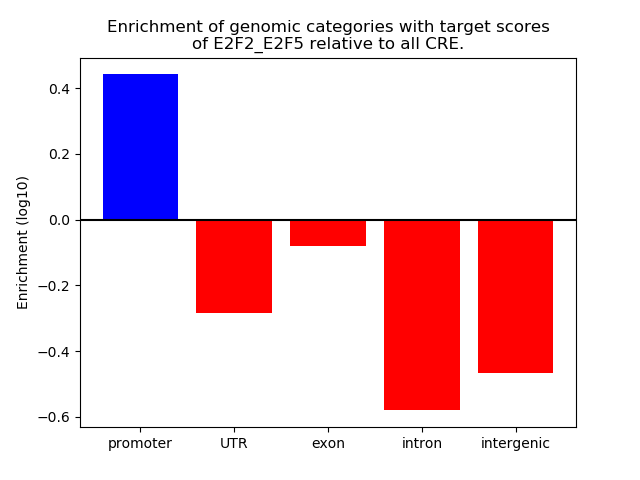
Activity of the E2F2_E2F5 motif across conditions
Conditions sorted by the z-value of the E2F2_E2F5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
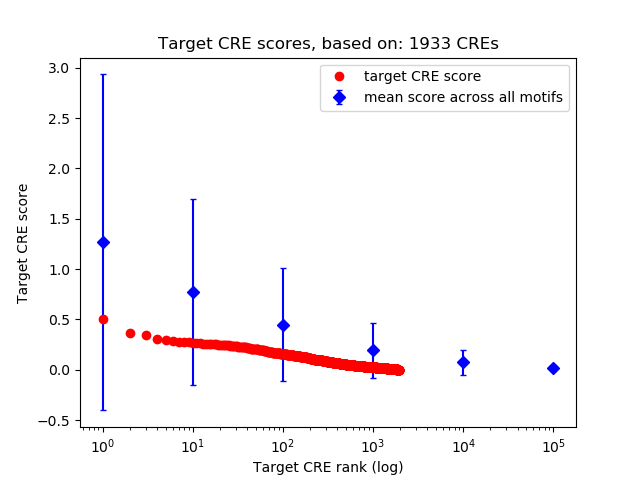
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_46755525_46755771 | 0.51 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
811 |
0.58 |
| chr5_130883505_130884015 | 0.37 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
15034 |
0.29 |
| chr1_184723746_184723985 | 0.35 |
EDEM3 |
ER degradation enhancer, mannosidase alpha-like 3 |
164 |
0.96 |
| chr6_27833013_27833577 | 0.30 |
HIST1H2AL |
histone cluster 1, H2al |
261 |
0.76 |
| chr4_89884993_89885181 | 0.30 |
FAM13A |
family with sequence similarity 13, member A |
65964 |
0.13 |
| chr18_19279350_19279501 | 0.29 |
ABHD3 |
abhydrolase domain containing 3 |
4224 |
0.13 |
| chr12_122984366_122984751 | 0.28 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
237 |
0.91 |
| chr3_119396008_119396222 | 0.27 |
COX17 |
COX17 cytochrome c oxidase copper chaperone |
78 |
0.97 |
| chr10_43951098_43951286 | 0.27 |
ENSG00000252532 |
. |
2321 |
0.26 |
| chr4_83812571_83812986 | 0.27 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
359 |
0.84 |
| chr2_136634130_136634399 | 0.27 |
MCM6 |
minichromosome maintenance complex component 6 |
268 |
0.91 |
| chr2_128174025_128174278 | 0.27 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
1852 |
0.28 |
| chr19_10305607_10305758 | 0.26 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
73 |
0.95 |
| chr10_12306595_12306746 | 0.26 |
ENSG00000265824 |
. |
3000 |
0.21 |
| chr16_56743265_56743416 | 0.26 |
RP11-343H19.1 |
|
20540 |
0.08 |
| chr12_12877828_12878109 | 0.26 |
APOLD1 |
apolipoprotein L domain containing 1 |
897 |
0.38 |
| chr17_80273991_80274142 | 0.25 |
CD7 |
CD7 molecule |
1362 |
0.29 |
| chr3_50487862_50488065 | 0.25 |
ENSG00000202322 |
. |
30751 |
0.11 |
| chr6_42858682_42858971 | 0.25 |
C6orf226 |
chromosome 6 open reading frame 226 |
272 |
0.86 |
| chr5_14582511_14582672 | 0.25 |
FAM105A |
family with sequence similarity 105, member A |
707 |
0.77 |
| chr1_23885059_23885312 | 0.25 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
1100 |
0.5 |
| chr5_39074216_39074429 | 0.25 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
169 |
0.97 |
| chrX_39966948_39967099 | 0.24 |
BCOR |
BCL6 corepressor |
10367 |
0.31 |
| chr1_205290578_205290810 | 0.24 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
189 |
0.94 |
| chr11_44748747_44748903 | 0.24 |
TSPAN18 |
tetraspanin 18 |
101 |
0.98 |
| chr19_37019294_37019507 | 0.24 |
ZNF260 |
zinc finger protein 260 |
162 |
0.51 |
| chr14_51706549_51708116 | 0.24 |
TMX1 |
thioredoxin-related transmembrane protein 1 |
446 |
0.81 |
| chrX_73164212_73165045 | 0.24 |
RP13-216E22.5 |
|
24692 |
0.23 |
| chr3_37035014_37035258 | 0.23 |
MLH1 |
mutL homolog 1 |
72 |
0.81 |
| chr3_85442393_85442544 | 0.23 |
ENSG00000264084 |
. |
7608 |
0.34 |
| chr2_677225_677432 | 0.23 |
TMEM18 |
transmembrane protein 18 |
111 |
0.54 |
| chr17_80709945_80710208 | 0.23 |
TBCD |
tubulin folding cofactor D |
42 |
0.95 |
| chr17_78120589_78120769 | 0.23 |
EIF4A3 |
eukaryotic translation initiation factor 4A3 |
259 |
0.9 |
| chr3_14989325_14989581 | 0.23 |
FGD5-AS1 |
FGD5 antisense RNA 1 |
53 |
0.72 |
| chrX_119619280_119620483 | 0.23 |
LAMP2 |
lysosomal-associated membrane protein 2 |
16661 |
0.2 |
| chr20_20693389_20693558 | 0.22 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
342 |
0.92 |
| chr19_50354007_50354329 | 0.22 |
PTOV1 |
prostate tumor overexpressed 1 |
30 |
0.62 |
| chr19_1438420_1438608 | 0.22 |
RPS15 |
ribosomal protein S15 |
95 |
0.9 |
| chr13_34392298_34392510 | 0.22 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
198 |
0.96 |
| chrX_102531488_102531657 | 0.22 |
TCEAL5 |
transcription elongation factor A (SII)-like 5 |
228 |
0.92 |
| chr17_58678123_58678617 | 0.22 |
PPM1D |
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
826 |
0.61 |
| chr16_10837539_10837690 | 0.21 |
NUBP1 |
nucleotide binding protein 1 |
29 |
0.98 |
| chrY_15815919_15816489 | 0.21 |
TMSB4Y |
thymosin beta 4, Y-linked |
757 |
0.72 |
| chr3_42814035_42814186 | 0.21 |
CCDC13 |
coiled-coil domain containing 13 |
635 |
0.55 |
| chr4_129731266_129731814 | 0.21 |
JADE1 |
jade family PHD finger 1 |
354 |
0.92 |
| chr8_90995843_90996298 | 0.21 |
NBN |
nibrin |
395 |
0.87 |
| chr21_46824152_46824303 | 0.21 |
COL18A1 |
collagen, type XVIII, alpha 1 |
825 |
0.56 |
| chr21_34697352_34697672 | 0.21 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
234 |
0.93 |
| chr5_42952303_42952454 | 0.21 |
SEPP1 |
selenoprotein P, plasma, 1 |
64884 |
0.09 |
| chr20_2082221_2082677 | 0.21 |
STK35 |
serine/threonine kinase 35 |
79 |
0.98 |
| chr12_14518654_14518864 | 0.20 |
ATF7IP |
activating transcription factor 7 interacting protein |
96 |
0.98 |
| chr11_46721727_46722111 | 0.20 |
ARHGAP1 |
Rho GTPase activating protein 1 |
230 |
0.69 |
| chr5_70196845_70197188 | 0.20 |
SERF1A |
small EDRK-rich factor 1A (telomeric) |
195 |
0.96 |
| chr17_66288424_66288865 | 0.20 |
ARSG |
arylsulfatase G |
985 |
0.41 |
| chr7_101459208_101459407 | 0.20 |
CUX1 |
cut-like homeobox 1 |
6 |
0.98 |
| chr3_31573575_31573846 | 0.20 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
572 |
0.85 |
| chr19_56146129_56146280 | 0.20 |
ZNF580 |
zinc finger protein 580 |
178 |
0.77 |
| chr19_17622493_17622725 | 0.20 |
PGLS |
6-phosphogluconolactonase |
65 |
0.94 |
| chr6_26233316_26233497 | 0.20 |
HIST1H1D |
histone cluster 1, H1d |
1810 |
0.12 |
| chr1_63249822_63250184 | 0.19 |
ATG4C |
autophagy related 4C, cysteine peptidase |
197 |
0.96 |
| chr1_222988010_222988273 | 0.19 |
DISP1 |
dispatched homolog 1 (Drosophila) |
57472 |
0.11 |
| chr6_27775872_27776194 | 0.19 |
HIST1H2AI |
histone cluster 1, H2ai |
134 |
0.69 |
| chr7_100034295_100034484 | 0.19 |
PPP1R35 |
protein phosphatase 1, regulatory subunit 35 |
201 |
0.72 |
| chr22_31615175_31615326 | 0.19 |
ENSG00000199695 |
. |
3589 |
0.13 |
| chr17_73975299_73975497 | 0.19 |
TEN1 |
TEN1 CST complex subunit |
46 |
0.49 |
| chr3_142838592_142838743 | 0.18 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
494 |
0.84 |
| chr16_84681720_84681934 | 0.18 |
KLHL36 |
kelch-like family member 36 |
304 |
0.9 |
| chr14_22961814_22961965 | 0.18 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
5718 |
0.11 |
| chr7_30634096_30634338 | 0.18 |
GARS |
glycyl-tRNA synthetase |
80 |
0.61 |
| chr15_57211218_57211369 | 0.18 |
TCF12 |
transcription factor 12 |
53 |
0.9 |
| chr9_98080006_98080370 | 0.18 |
FANCC |
Fanconi anemia, complementation group C |
204 |
0.96 |
| chr10_99051898_99052365 | 0.18 |
ARHGAP19-SLIT1 |
ARHGAP19-SLIT1 readthrough (NMD candidate) |
253 |
0.48 |
| chr14_97879221_97879798 | 0.18 |
ENSG00000240730 |
. |
117001 |
0.07 |
| chr6_52149357_52149559 | 0.17 |
MCM3 |
minichromosome maintenance complex component 3 |
47 |
0.98 |
| chr6_31619842_31620065 | 0.17 |
BAG6 |
BCL2-associated athanogene 6 |
18 |
0.71 |
| chr1_149982780_149982931 | 0.17 |
OTUD7B |
OTU domain containing 7B |
230 |
0.91 |
| chr2_160569063_160569519 | 0.17 |
MARCH7 |
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
273 |
0.94 |
| chr7_7679470_7679820 | 0.17 |
RPA3 |
replication protein A3, 14kDa |
12 |
0.99 |
| chr1_12201087_12201238 | 0.17 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
15206 |
0.15 |
| chr1_23857954_23858302 | 0.17 |
E2F2 |
E2F transcription factor 2 |
416 |
0.83 |
| chr6_142467968_142468269 | 0.17 |
VTA1 |
vesicle (multivesicular body) trafficking 1 |
249 |
0.93 |
| chr6_36164322_36164500 | 0.17 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
110 |
0.88 |
| chr15_41952602_41953001 | 0.17 |
MGA |
MGA, MAX dimerization protein |
191 |
0.94 |
| chr17_10600647_10600878 | 0.17 |
SCO1 |
SCO1 cytochrome c oxidase assembly protein |
85 |
0.59 |
| chrX_123095829_123096146 | 0.17 |
STAG2 |
stromal antigen 2 |
43 |
0.98 |
| chr5_69321431_69321977 | 0.17 |
SERF1B |
small EDRK-rich factor 1B (centromeric) |
343 |
0.9 |
| chr15_34634257_34634507 | 0.17 |
NOP10 |
NOP10 ribonucleoprotein |
942 |
0.33 |
| chr2_242625574_242625851 | 0.17 |
DTYMK |
deoxythymidylate kinase (thymidylate kinase) |
415 |
0.72 |
| chr8_38854817_38854968 | 0.17 |
ADAM9 |
ADAM metallopeptidase domain 9 |
387 |
0.57 |
| chr11_12132484_12132785 | 0.17 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
496 |
0.87 |
| chr19_45418370_45418582 | 0.17 |
APOC1 |
apolipoprotein C-I |
350 |
0.72 |
| chr8_18973333_18973484 | 0.16 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
31168 |
0.23 |
| chr21_38630680_38631091 | 0.16 |
DSCR3 |
Down syndrome critical region gene 3 |
8738 |
0.17 |
| chr21_44582259_44582410 | 0.16 |
AP001631.10 |
|
53 |
0.97 |
| chr19_1407342_1407493 | 0.16 |
DAZAP1 |
DAZ associated protein 1 |
151 |
0.88 |
| chr10_63809082_63810748 | 0.16 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
945 |
0.69 |
| chr8_71971447_71971598 | 0.16 |
RP11-326E22.1 |
|
96170 |
0.09 |
| chr3_105088175_105088464 | 0.16 |
ALCAM |
activated leukocyte cell adhesion molecule |
2132 |
0.49 |
| chrX_40027620_40027771 | 0.16 |
BCOR |
BCL6 corepressor |
8878 |
0.31 |
| chr13_108870830_108871120 | 0.16 |
ABHD13 |
abhydrolase domain containing 13 |
248 |
0.5 |
| chr17_44344409_44344766 | 0.16 |
RP11-259G18.1 |
|
191 |
0.93 |
| chr19_46144786_46145014 | 0.16 |
C19orf83 |
chromosome 19 open reading frame 83 |
49 |
0.91 |
| chr19_11616618_11616769 | 0.15 |
ZNF653 |
zinc finger protein 653 |
45 |
0.95 |
| chr1_120612553_120612848 | 0.15 |
NOTCH2 |
notch 2 |
460 |
0.87 |
| chr1_235247521_235247672 | 0.15 |
ENSG00000207181 |
. |
43656 |
0.13 |
| chr14_60982037_60982188 | 0.15 |
C14orf39 |
chromosome 14 open reading frame 39 |
149 |
0.67 |
| chr10_7449615_7449918 | 0.15 |
SFMBT2 |
Scm-like with four mbt domains 2 |
941 |
0.7 |
| chr13_45694691_45695194 | 0.15 |
GTF2F2 |
general transcription factor IIF, polypeptide 2, 30kDa |
292 |
0.87 |
| chr5_178053297_178053654 | 0.15 |
CLK4 |
CDC-like kinase 4 |
556 |
0.76 |
| chr3_69100609_69100945 | 0.15 |
TMF1 |
TATA element modulatory factor 1 |
677 |
0.58 |
| chr19_16295903_16296067 | 0.15 |
FAM32A |
family with sequence similarity 32, member A |
206 |
0.9 |
| chr12_2986635_2986949 | 0.15 |
RHNO1 |
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
403 |
0.54 |
| chr12_31811913_31812109 | 0.15 |
METTL20 |
methyltransferase like 20 |
110 |
0.96 |
| chr1_28574021_28574215 | 0.15 |
RP5-1092A3.4 |
|
6154 |
0.12 |
| chr19_633126_633363 | 0.15 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
353 |
0.74 |
| chr14_81687775_81688125 | 0.15 |
GTF2A1 |
general transcription factor IIA, 1, 19/37kDa |
375 |
0.88 |
| chr5_74062328_74062877 | 0.15 |
NSA2 |
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
215 |
0.57 |
| chr6_32098148_32098328 | 0.15 |
FKBPL |
FK506 binding protein like |
170 |
0.87 |
| chr5_43064729_43065087 | 0.15 |
CTD-2201E18.3 |
|
2613 |
0.21 |
| chr4_100870352_100870894 | 0.15 |
RP11-15B17.1 |
|
540 |
0.59 |
| chr2_175350503_175351393 | 0.15 |
GPR155 |
G protein-coupled receptor 155 |
817 |
0.65 |
| chr4_1161098_1161472 | 0.15 |
RP11-20I20.4 |
|
281 |
0.86 |
| chr10_22904775_22905136 | 0.15 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
24313 |
0.26 |
| chr6_3229020_3229250 | 0.15 |
TUBB2B |
tubulin, beta 2B class IIb |
1166 |
0.44 |
| chr17_4607485_4607753 | 0.14 |
PELP1 |
proline, glutamate and leucine rich protein 1 |
1 |
0.56 |
| chr1_36234823_36235223 | 0.14 |
CLSPN |
claspin |
506 |
0.76 |
| chr8_23021171_23021491 | 0.14 |
TNFRSF10D |
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
212 |
0.91 |
| chr8_22551556_22552015 | 0.14 |
EGR3 |
early growth response 3 |
970 |
0.44 |
| chr15_44719862_44720599 | 0.14 |
CTDSPL2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
218 |
0.85 |
| chr15_41047576_41047750 | 0.14 |
RMDN3 |
regulator of microtubule dynamics 3 |
129 |
0.93 |
| chr5_180649554_180650164 | 0.14 |
ENSG00000264732 |
. |
226 |
0.59 |
| chr4_39045572_39046195 | 0.14 |
KLHL5 |
kelch-like family member 5 |
776 |
0.67 |
| chr4_84376358_84376962 | 0.14 |
HELQ |
helicase, POLQ-like |
316 |
0.61 |
| chr7_39991401_39992022 | 0.14 |
CDK13 |
cyclin-dependent kinase 13 |
1602 |
0.37 |
| chr11_34937157_34937669 | 0.14 |
PDHX |
pyruvate dehydrogenase complex, component X |
37 |
0.89 |
| chr5_132083512_132083663 | 0.14 |
CCNI2 |
cyclin I family, member 2 |
450 |
0.75 |
| chr2_173293363_173293761 | 0.14 |
AC078883.4 |
|
231 |
0.88 |
| chr11_22215104_22215474 | 0.14 |
ANO5 |
anoctamin 5 |
567 |
0.84 |
| chr9_139433380_139433586 | 0.14 |
RP11-413M3.4 |
|
3850 |
0.11 |
| chr20_18269271_18269509 | 0.14 |
ZNF133 |
zinc finger protein 133 |
212 |
0.9 |
| chr3_197476356_197476565 | 0.14 |
FYTTD1 |
forty-two-three domain containing 1 |
36 |
0.58 |
| chrX_45709671_45710004 | 0.14 |
ENSG00000212347 |
. |
42577 |
0.19 |
| chr11_77850351_77850689 | 0.14 |
ALG8 |
ALG8, alpha-1,3-glucosyltransferase |
118 |
0.72 |
| chr6_27099516_27099912 | 0.14 |
HIST1H2BJ |
histone cluster 1, H2bj |
613 |
0.5 |
| chr9_99180680_99180947 | 0.14 |
ZNF367 |
zinc finger protein 367 |
202 |
0.95 |
| chr11_121322497_121322843 | 0.14 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
242 |
0.88 |
| chr6_82460724_82461189 | 0.14 |
FAM46A |
family with sequence similarity 46, member A |
514 |
0.8 |
| chr9_134152676_134153362 | 0.13 |
FAM78A |
family with sequence similarity 78, member A |
1085 |
0.5 |
| chr14_55877782_55878514 | 0.13 |
ATG14 |
autophagy related 14 |
428 |
0.86 |
| chrY_15863423_15864046 | 0.13 |
KALP |
Kallmann syndrome sequence pseudogene |
198 |
0.96 |
| chr5_169064267_169064974 | 0.13 |
DOCK2 |
dedicator of cytokinesis 2 |
369 |
0.91 |
| chr6_84568793_84569179 | 0.13 |
CYB5R4 |
cytochrome b5 reductase 4 |
376 |
0.89 |
| chr5_79553424_79553684 | 0.13 |
SERINC5 |
serine incorporator 5 |
1656 |
0.36 |
| chr11_62622072_62622332 | 0.13 |
ENSG00000255717 |
. |
35 |
0.92 |
| chr12_83080910_83081086 | 0.13 |
TMTC2 |
transmembrane and tetratricopeptide repeat containing 2 |
218 |
0.97 |
| chr12_27398115_27398400 | 0.13 |
STK38L |
serine/threonine kinase 38 like |
333 |
0.92 |
| chr9_27529480_27529683 | 0.13 |
MOB3B |
MOB kinase activator 3B |
198 |
0.95 |
| chr21_34914612_34915092 | 0.13 |
SON |
SON DNA binding protein |
72 |
0.65 |
| chr16_29816076_29816359 | 0.13 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
1210 |
0.2 |
| chr17_56565203_56565440 | 0.13 |
HSF5 |
heat shock transcription factor family member 5 |
424 |
0.77 |
| chr3_187461693_187462048 | 0.13 |
BCL6 |
B-cell CLL/lymphoma 6 |
1645 |
0.42 |
| chr5_134094069_134094276 | 0.13 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
299 |
0.88 |
| chr5_131628993_131629263 | 0.13 |
SLC22A4 |
solute carrier family 22 (organic cation/zwitterion transporter), member 4 |
1008 |
0.42 |
| chr8_73921692_73921901 | 0.13 |
TERF1 |
telomeric repeat binding factor (NIMA-interacting) 1 |
684 |
0.67 |
| chr19_3812640_3812791 | 0.13 |
MATK |
megakaryocyte-associated tyrosine kinase |
10588 |
0.11 |
| chr14_60716102_60716311 | 0.13 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
47 |
0.94 |
| chr9_70402575_70402838 | 0.13 |
IGKV1OR-3 |
immunoglobulin kappa variable 1/OR-3 (pseudogene) |
7616 |
0.24 |
| chr15_43414561_43415376 | 0.13 |
TMEM62 |
transmembrane protein 62 |
509 |
0.77 |
| chr18_11850148_11850299 | 0.13 |
CHMP1B |
charged multivesicular body protein 1B |
1172 |
0.34 |
| chr6_99281230_99281408 | 0.13 |
POU3F2 |
POU class 3 homeobox 2 |
1261 |
0.63 |
| chr13_49105975_49106823 | 0.13 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
814 |
0.75 |
| chrX_106871756_106872595 | 0.13 |
PRPS1 |
phosphoribosyl pyrophosphate synthetase 1 |
382 |
0.9 |
| chr13_49821689_49822053 | 0.13 |
CDADC1 |
cytidine and dCMP deaminase domain containing 1 |
176 |
0.96 |
| chr12_57481229_57481548 | 0.12 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
1289 |
0.28 |
| chr1_231377063_231377587 | 0.12 |
GNPAT |
glyceronephosphate O-acyltransferase |
363 |
0.5 |
| chr10_85954746_85954956 | 0.12 |
CDHR1 |
cadherin-related family member 1 |
437 |
0.79 |
| chr2_109336300_109336755 | 0.12 |
RANBP2 |
RAN binding protein 2 |
590 |
0.78 |
| chr20_25387997_25388359 | 0.12 |
GINS1 |
GINS complex subunit 1 (Psf1 homolog) |
185 |
0.95 |
| chr6_150039328_150039894 | 0.12 |
LATS1 |
large tumor suppressor kinase 1 |
219 |
0.92 |
| chr5_72251895_72252188 | 0.12 |
FCHO2 |
FCH domain only 2 |
143 |
0.97 |
| chr1_154934578_154934948 | 0.12 |
PYGO2 |
pygopus family PHD finger 2 |
251 |
0.62 |
| chr20_23028362_23028696 | 0.12 |
THBD |
thrombomodulin |
1849 |
0.28 |
| chr17_6938780_6939115 | 0.12 |
SLC16A13 |
solute carrier family 16, member 13 |
447 |
0.6 |
| chr12_49783044_49783195 | 0.12 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
21789 |
0.13 |
| chr11_129242275_129242426 | 0.12 |
BARX2 |
BARX homeobox 2 |
3485 |
0.36 |
| chr11_65190939_65191303 | 0.12 |
ENSG00000245532 |
. |
20808 |
0.1 |
| chr1_54872617_54872800 | 0.12 |
SSBP3 |
single stranded DNA binding protein 3 |
616 |
0.78 |
| chr1_201562555_201562715 | 0.12 |
AC096677.1 |
Uncharacterized protein ENSP00000471857 |
29378 |
0.14 |
| chr2_85107193_85107344 | 0.12 |
TRABD2A |
TraB domain containing 2A |
938 |
0.57 |
| chr3_45729758_45730513 | 0.12 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
239 |
0.78 |
| chr1_205180919_205181105 | 0.12 |
DSTYK |
dual serine/threonine and tyrosine protein kinase |
285 |
0.9 |
| chr7_19748667_19748818 | 0.12 |
TWISTNB |
TWIST neighbor |
32 |
0.98 |
| chr5_112315292_112315678 | 0.12 |
DCP2 |
decapping mRNA 2 |
3006 |
0.27 |
| chr11_67188360_67188666 | 0.12 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
139 |
0.88 |
| chr1_28241326_28241544 | 0.12 |
RPA2 |
replication protein A2, 32kDa |
178 |
0.92 |
| chr10_70320968_70321193 | 0.12 |
TET1 |
tet methylcytosine dioxygenase 1 |
667 |
0.7 |
| chr1_151118477_151118817 | 0.12 |
SEMA6C |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
286 |
0.8 |
| chr6_106968131_106968391 | 0.12 |
AIM1 |
absent in melanoma 1 |
8531 |
0.21 |
| chr22_38901190_38901707 | 0.12 |
DDX17 |
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
869 |
0.54 |
| chr1_3568811_3568962 | 0.11 |
TP73 |
tumor protein p73 |
198 |
0.72 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.3 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0044818 | mitotic DNA replication checkpoint(GO:0033314) mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:2000104 | negative regulation of DNA-dependent DNA replication(GO:2000104) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.7 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.0 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.3 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0034445 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.0 | REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | Genes involved in Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |