Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
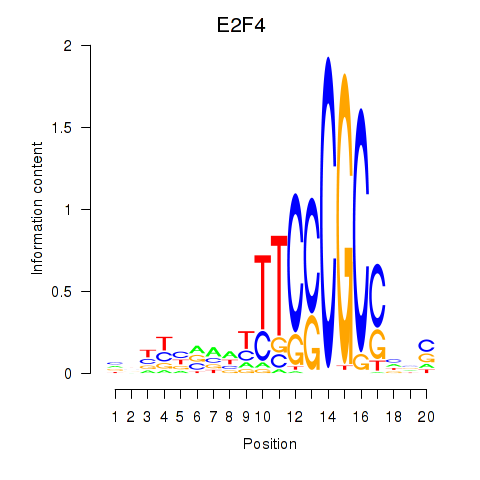
Results for E2F4
Z-value: 0.84
Transcription factors associated with E2F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F4
|
ENSG00000205250.4 | E2F transcription factor 4 |
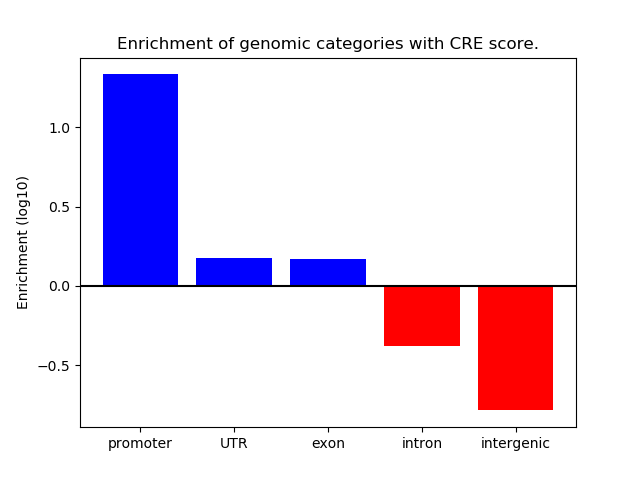
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_67227932_67228083 | E2F4 | 848 | 0.306548 | 0.16 | 6.8e-01 | Click! |
| chr16_67226442_67227225 | E2F4 | 761 | 0.343028 | -0.13 | 7.4e-01 | Click! |
| chr16_67226195_67226432 | E2F4 | 241 | 0.774486 | 0.08 | 8.4e-01 | Click! |
| chr16_67225776_67226012 | E2F4 | 178 | 0.831564 | 0.01 | 9.8e-01 | Click! |
Activity of the E2F4 motif across conditions
Conditions sorted by the z-value of the E2F4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
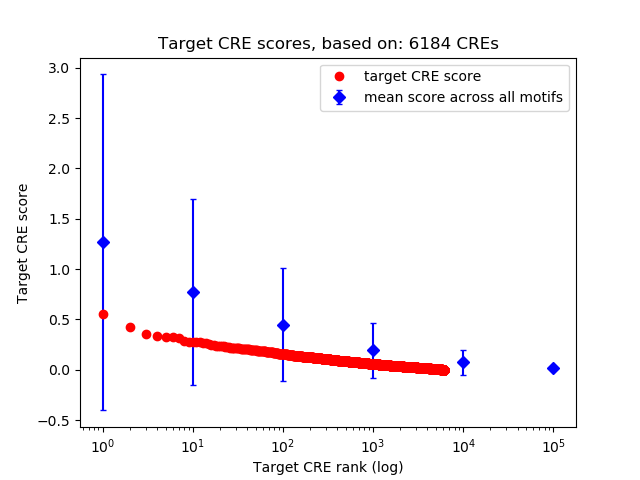
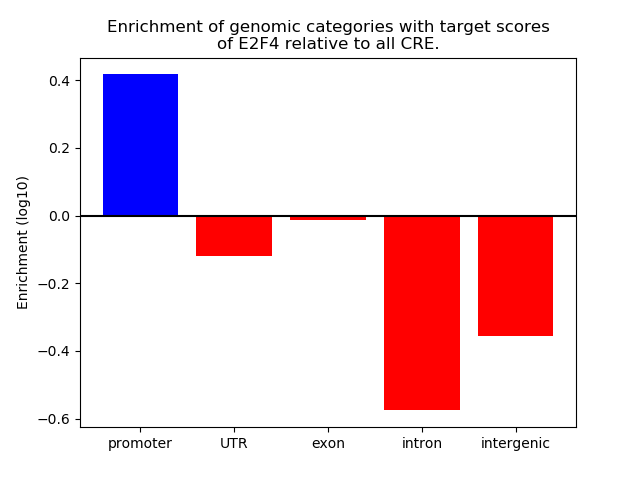
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_99729815_99730045 | 0.56 |
AL109767.1 |
|
645 |
0.75 |
| chr1_91966366_91967474 | 0.42 |
CDC7 |
cell division cycle 7 |
225 |
0.96 |
| chr1_111888607_111889201 | 0.36 |
PIFO |
primary cilia formation |
6 |
0.97 |
| chr17_35860978_35861129 | 0.33 |
DUSP14 |
dual specificity phosphatase 14 |
8928 |
0.19 |
| chr8_1430658_1430809 | 0.33 |
DLGAP2 |
discs, large (Drosophila) homolog-associated protein 2 |
18799 |
0.28 |
| chr6_391121_391497 | 0.32 |
IRF4 |
interferon regulatory factor 4 |
430 |
0.9 |
| chr17_37762810_37762961 | 0.32 |
NEUROD2 |
neuronal differentiation 2 |
1311 |
0.34 |
| chr2_202098878_202099103 | 0.28 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
761 |
0.65 |
| chrX_68047832_68048201 | 0.28 |
EFNB1 |
ephrin-B1 |
824 |
0.77 |
| chrX_106448742_106449790 | 0.28 |
NUP62CL |
nucleoporin 62kDa C-terminal like |
397 |
0.61 |
| chr3_171176844_171177618 | 0.28 |
TNIK |
TRAF2 and NCK interacting kinase |
621 |
0.77 |
| chr13_34116896_34117047 | 0.27 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
192204 |
0.03 |
| chr16_86542425_86542576 | 0.27 |
FOXF1 |
forkhead box F1 |
1633 |
0.41 |
| chr20_2451178_2451431 | 0.27 |
SNRPB |
small nuclear ribonucleoprotein polypeptides B and B1 |
127 |
0.95 |
| chr1_212780870_212781021 | 0.26 |
ATF3 |
activating transcription factor 3 |
1067 |
0.45 |
| chr17_73105615_73105966 | 0.25 |
ARMC7 |
armadillo repeat containing 7 |
257 |
0.82 |
| chr1_234040313_234040583 | 0.24 |
SLC35F3 |
solute carrier family 35, member F3 |
231 |
0.96 |
| chr4_149364707_149365251 | 0.24 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
871 |
0.75 |
| chr5_138728866_138729168 | 0.24 |
PROB1 |
proline-rich basic protein 1 |
1868 |
0.18 |
| chr19_49931395_49931706 | 0.24 |
GFY |
golgi-associated, olfactory signaling regulator |
4544 |
0.07 |
| chr13_32889728_32890299 | 0.24 |
BRCA2 |
breast cancer 2, early onset |
371 |
0.59 |
| chr15_41199252_41199466 | 0.23 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
12694 |
0.11 |
| chr19_56014942_56015267 | 0.23 |
SSC5D |
scavenger receptor cysteine rich domain containing (5 domains) |
9846 |
0.08 |
| chr10_79050334_79050485 | 0.22 |
RP11-328K22.1 |
|
23290 |
0.23 |
| chr8_41624406_41625533 | 0.22 |
ANK1 |
ankyrin 1, erythrocytic |
30171 |
0.15 |
| chr1_155162943_155163205 | 0.22 |
MUC1 |
mucin 1, cell surface associated |
368 |
0.6 |
| chr12_69201397_69201692 | 0.22 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
412 |
0.8 |
| chr3_51720796_51721049 | 0.22 |
ENSG00000201595 |
. |
7676 |
0.15 |
| chr11_62367040_62367191 | 0.22 |
MTA2 |
metastasis associated 1 family, member 2 |
326 |
0.7 |
| chr20_30467916_30468108 | 0.22 |
TTLL9 |
tubulin tyrosine ligase-like family, member 9 |
380 |
0.8 |
| chr16_68679557_68679708 | 0.22 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
428 |
0.45 |
| chr17_3658508_3658859 | 0.21 |
ITGAE |
integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
11849 |
0.12 |
| chr17_35293190_35293341 | 0.21 |
RP11-445F12.1 |
|
656 |
0.45 |
| chrY_16636064_16636659 | 0.21 |
NLGN4Y |
neuroligin 4, Y-linked |
93 |
0.99 |
| chr2_128785898_128786120 | 0.21 |
SAP130 |
Sin3A-associated protein, 130kDa |
315 |
0.91 |
| chr17_38024211_38024362 | 0.21 |
ZPBP2 |
zona pellucida binding protein 2 |
131 |
0.95 |
| chr17_59940229_59940941 | 0.21 |
BRIP1 |
BRCA1 interacting protein C-terminal helicase 1 |
120 |
0.97 |
| chr19_50249421_50249636 | 0.21 |
TSKS |
testis-specific serine kinase substrate |
1107 |
0.28 |
| chrX_73755392_73756724 | 0.20 |
SLC16A2 |
solute carrier family 16, member 2 (thyroid hormone transporter) |
15103 |
0.23 |
| chr11_34379895_34380202 | 0.20 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
493 |
0.86 |
| chr11_726415_726634 | 0.20 |
AP006621.9 |
|
523 |
0.57 |
| chr2_10184175_10184326 | 0.20 |
KLF11 |
Kruppel-like factor 11 |
52 |
0.96 |
| chr20_2820909_2821269 | 0.20 |
PCED1A |
PC-esterase domain containing 1A |
228 |
0.52 |
| chr13_22248486_22248637 | 0.20 |
FGF9 |
fibroblast growth factor 9 |
3039 |
0.33 |
| chr17_70118144_70118357 | 0.20 |
SOX9 |
SRY (sex determining region Y)-box 9 |
1089 |
0.67 |
| chr1_47899465_47899709 | 0.20 |
FOXD2-AS1 |
FOXD2 antisense RNA 1 (head to head) |
726 |
0.57 |
| chr11_1567498_1567649 | 0.19 |
DUSP8 |
dual specificity phosphatase 8 |
19593 |
0.12 |
| chr1_6550194_6550563 | 0.19 |
PLEKHG5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
252 |
0.87 |
| chr20_62272433_62273064 | 0.19 |
STMN3 |
stathmin-like 3 |
11371 |
0.09 |
| chr19_38701881_38702032 | 0.19 |
ENSG00000266428 |
. |
11082 |
0.1 |
| chr17_58469595_58469783 | 0.19 |
USP32 |
ubiquitin specific peptidase 32 |
80 |
0.53 |
| chr5_140997925_140998467 | 0.19 |
AC008781.7 |
|
215 |
0.56 |
| chr14_92588238_92588992 | 0.19 |
CPSF2 |
cleavage and polyadenylation specific factor 2, 100kDa |
299 |
0.54 |
| chr1_26947517_26947992 | 0.19 |
ENSG00000238316 |
. |
21126 |
0.12 |
| chrX_70837224_70837375 | 0.19 |
CXCR3 |
chemokine (C-X-C motif) receptor 3 |
1061 |
0.51 |
| chr2_157198349_157198757 | 0.19 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
307 |
0.93 |
| chr1_4716023_4716174 | 0.19 |
AJAP1 |
adherens junctions associated protein 1 |
993 |
0.73 |
| chrY_15815919_15816489 | 0.19 |
TMSB4Y |
thymosin beta 4, Y-linked |
757 |
0.72 |
| chr14_94254776_94255139 | 0.18 |
PRIMA1 |
proline rich membrane anchor 1 |
130 |
0.98 |
| chrX_1780511_1780662 | 0.18 |
ASMT |
acetylserotonin O-methyltransferase |
25295 |
0.22 |
| chr17_19015403_19015696 | 0.18 |
ENSG00000262202 |
. |
400 |
0.71 |
| chr16_68270253_68270605 | 0.18 |
ESRP2 |
epithelial splicing regulatory protein 2 |
58 |
0.91 |
| chr11_777311_777493 | 0.18 |
PDDC1 |
Parkinson disease 7 domain containing 1 |
69 |
0.63 |
| chr17_35165532_35165707 | 0.18 |
RP11-445F12.1 |
|
128302 |
0.04 |
| chr7_100202948_100203247 | 0.18 |
PCOLCE-AS1 |
PCOLCE antisense RNA 1 |
1268 |
0.23 |
| chrY_1730510_1730661 | 0.18 |
ENSG00000251841 |
. |
922205 |
0.0 |
| chr7_560180_560411 | 0.18 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
362 |
0.88 |
| chr19_639517_639685 | 0.18 |
FGF22 |
fibroblast growth factor 22 |
294 |
0.79 |
| chr17_38020283_38020945 | 0.18 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
173 |
0.93 |
| chr16_30081137_30081369 | 0.18 |
ALDOA |
aldolase A, fructose-bisphosphate |
354 |
0.71 |
| chr1_161088064_161088385 | 0.18 |
NIT1 |
nitrilase 1 |
316 |
0.43 |
| chr18_28621917_28622068 | 0.18 |
DSC3 |
desmocollin 3 |
715 |
0.67 |
| chr11_2812990_2813414 | 0.18 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
69596 |
0.08 |
| chr5_114504926_114505203 | 0.18 |
TRIM36 |
tripartite motif containing 36 |
542 |
0.77 |
| chr19_46282643_46282874 | 0.17 |
DMPK |
dystrophia myotonica-protein kinase |
1041 |
0.28 |
| chr20_21086226_21086377 | 0.17 |
ENSG00000199509 |
. |
33414 |
0.2 |
| chr7_134330596_134330773 | 0.17 |
BPGM |
2,3-bisphosphoglycerate mutase |
876 |
0.7 |
| chr10_124909262_124909943 | 0.17 |
HMX2 |
H6 family homeobox 2 |
1964 |
0.29 |
| chr16_2317895_2318079 | 0.17 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
68 |
0.53 |
| chr4_159592552_159593087 | 0.17 |
C4orf46 |
chromosome 4 open reading frame 46 |
225 |
0.72 |
| chr17_7791100_7791439 | 0.17 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
840 |
0.38 |
| chr4_154680740_154681409 | 0.17 |
RNF175 |
ring finger protein 175 |
75 |
0.97 |
| chr17_36667183_36667334 | 0.17 |
ARHGAP23 |
Rho GTPase activating protein 23 |
13223 |
0.16 |
| chr10_70090873_70091608 | 0.17 |
HNRNPH3 |
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
592 |
0.65 |
| chr14_99712277_99712758 | 0.17 |
AL109767.1 |
|
16768 |
0.21 |
| chr19_49630930_49631277 | 0.17 |
PPFIA3 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
28 |
0.94 |
| chr1_24513240_24513925 | 0.17 |
IFNLR1 |
interferon, lambda receptor 1 |
156 |
0.95 |
| chr5_109025309_109025497 | 0.17 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
336 |
0.89 |
| chr1_11741356_11741653 | 0.16 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
233 |
0.89 |
| chr11_111307398_111307568 | 0.16 |
RP11-794P6.3 |
|
13110 |
0.11 |
| chr19_4584029_4584180 | 0.16 |
SEMA6B |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
24284 |
0.09 |
| chr1_15500593_15500744 | 0.16 |
C1orf195 |
chromosome 1 open reading frame 195 |
2855 |
0.29 |
| chr1_228603625_228604274 | 0.16 |
TRIM17 |
tripartite motif containing 17 |
219 |
0.85 |
| chr19_35610158_35610309 | 0.16 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
34 |
0.95 |
| chr10_22726004_22726972 | 0.16 |
RP11-301N24.3 |
|
76387 |
0.09 |
| chr14_99740559_99741090 | 0.16 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
2963 |
0.29 |
| chr2_25537366_25537517 | 0.16 |
ENSG00000221445 |
. |
14149 |
0.21 |
| chr14_91883398_91883667 | 0.16 |
CCDC88C |
coiled-coil domain containing 88C |
158 |
0.97 |
| chr21_43513996_43514147 | 0.16 |
C21orf128 |
chromosome 21 open reading frame 128 |
14573 |
0.18 |
| chr4_84255463_84255632 | 0.16 |
HPSE |
heparanase |
388 |
0.89 |
| chr1_1241228_1241379 | 0.16 |
ACAP3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
1966 |
0.13 |
| chr5_156887380_156887596 | 0.16 |
CTB-109A12.1 |
|
402 |
0.51 |
| chr18_33767527_33768072 | 0.16 |
MOCOS |
molybdenum cofactor sulfurase |
317 |
0.55 |
| chr11_64636454_64636798 | 0.16 |
EHD1 |
EH-domain containing 1 |
6512 |
0.1 |
| chr19_50935416_50935771 | 0.15 |
MYBPC2 |
myosin binding protein C, fast type |
567 |
0.57 |
| chr8_135724729_135724908 | 0.15 |
ZFAT |
zinc finger and AT hook domain containing |
387 |
0.91 |
| chr10_6161799_6162068 | 0.15 |
RBM17 |
RNA binding motif protein 17 |
14999 |
0.14 |
| chr3_25706566_25706764 | 0.15 |
ENSG00000264219 |
. |
235 |
0.53 |
| chr19_49403222_49403463 | 0.15 |
NUCB1 |
nucleobindin 1 |
35 |
0.93 |
| chr10_7860903_7861171 | 0.15 |
TAF3 |
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
570 |
0.78 |
| chr1_227916358_227916819 | 0.15 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
230 |
0.92 |
| chr21_47743435_47743618 | 0.15 |
C21orf58 |
chromosome 21 open reading frame 58 |
245 |
0.71 |
| chr9_131655035_131655246 | 0.15 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
10359 |
0.11 |
| chr11_119562021_119562172 | 0.15 |
ENSG00000199217 |
. |
35075 |
0.14 |
| chr3_134515339_134515664 | 0.15 |
EPHB1 |
EPH receptor B1 |
518 |
0.8 |
| chr6_33589286_33590289 | 0.15 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
626 |
0.65 |
| chr15_30484201_30484352 | 0.15 |
ENSG00000221785 |
. |
47196 |
0.09 |
| chr9_140210185_140210336 | 0.15 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
13557 |
0.09 |
| chr17_6927436_6927587 | 0.15 |
BCL6B |
B-cell CLL/lymphoma 6, member B |
715 |
0.38 |
| chr5_172067951_172068128 | 0.15 |
NEURL1B |
neuralized E3 ubiquitin protein ligase 1B |
230 |
0.92 |
| chr20_55205529_55205819 | 0.15 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
264 |
0.9 |
| chr19_50063668_50063821 | 0.15 |
NOSIP |
nitric oxide synthase interacting protein |
143 |
0.88 |
| chr6_52226992_52227834 | 0.15 |
PAQR8 |
progestin and adipoQ receptor family member VIII |
169 |
0.96 |
| chr1_20811158_20811808 | 0.15 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
1230 |
0.48 |
| chr9_100995758_100995909 | 0.14 |
TBC1D2 |
TBC1 domain family, member 2 |
20001 |
0.21 |
| chr10_92631760_92632056 | 0.14 |
RPP30 |
ribonuclease P/MRP 30kDa subunit |
168 |
0.95 |
| chr1_3388211_3388362 | 0.14 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
98 |
0.97 |
| chr22_39640196_39640514 | 0.14 |
PDGFB |
platelet-derived growth factor beta polypeptide |
401 |
0.8 |
| chr16_30365797_30366566 | 0.14 |
CD2BP2 |
CD2 (cytoplasmic tail) binding protein 2 |
74 |
0.93 |
| chr2_71192167_71192590 | 0.14 |
ENSG00000241159 |
. |
323 |
0.74 |
| chr1_107682797_107683388 | 0.14 |
NTNG1 |
netrin G1 |
350 |
0.93 |
| chr14_69094821_69095326 | 0.14 |
CTD-2325P2.4 |
|
89 |
0.98 |
| chr19_1470719_1471158 | 0.14 |
C19orf25 |
chromosome 19 open reading frame 25 |
7958 |
0.07 |
| chr11_3819249_3819602 | 0.14 |
PGAP2 |
post-GPI attachment to proteins 2 |
223 |
0.66 |
| chr2_43454043_43454388 | 0.14 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
467 |
0.83 |
| chr17_75136159_75136684 | 0.14 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
397 |
0.86 |
| chr8_22549992_22550198 | 0.14 |
EGR3 |
early growth response 3 |
168 |
0.92 |
| chr16_87160456_87160743 | 0.14 |
RP11-899L11.3 |
|
88922 |
0.08 |
| chr18_43417850_43418214 | 0.14 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
226 |
0.94 |
| chr11_2291181_2291518 | 0.14 |
ASCL2 |
achaete-scute family bHLH transcription factor 2 |
833 |
0.53 |
| chr16_79844626_79844777 | 0.14 |
ENSG00000221330 |
. |
141067 |
0.05 |
| chr20_55965895_55966111 | 0.14 |
RBM38 |
RNA binding motif protein 38 |
460 |
0.74 |
| chr3_30655645_30655796 | 0.14 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
7627 |
0.31 |
| chr19_56014768_56014919 | 0.14 |
SSC5D |
scavenger receptor cysteine rich domain containing (5 domains) |
9585 |
0.08 |
| chr17_66016231_66016439 | 0.14 |
KPNA2 |
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
15300 |
0.15 |
| chr10_23633268_23633917 | 0.14 |
C10orf67 |
chromosome 10 open reading frame 67 |
182 |
0.86 |
| chr7_25902294_25902669 | 0.14 |
ENSG00000199085 |
. |
87125 |
0.1 |
| chr1_175162248_175162570 | 0.14 |
KIAA0040 |
KIAA0040 |
330 |
0.93 |
| chr13_110439180_110439367 | 0.14 |
IRS2 |
insulin receptor substrate 2 |
358 |
0.92 |
| chrX_101410005_101410156 | 0.14 |
BEX5 |
brain expressed, X-linked 5 |
682 |
0.64 |
| chr12_113590786_113591379 | 0.14 |
CCDC42B |
coiled-coil domain containing 42B |
47 |
0.95 |
| chr18_24128407_24128977 | 0.14 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
192 |
0.96 |
| chr17_66561126_66561277 | 0.14 |
AC079210.1 |
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
21832 |
0.18 |
| chr17_14204490_14204770 | 0.14 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
230 |
0.95 |
| chr17_38720511_38720770 | 0.13 |
CCR7 |
chemokine (C-C motif) receptor 7 |
1071 |
0.49 |
| chr13_109147564_109147910 | 0.13 |
MYO16 |
myosin XVI |
100763 |
0.08 |
| chr17_79917712_79918256 | 0.13 |
NOTUM |
notum pectinacetylesterase homolog (Drosophila) |
298 |
0.72 |
| chr9_971958_972221 | 0.13 |
DMRT3 |
doublesex and mab-3 related transcription factor 3 |
4875 |
0.26 |
| chr12_103888876_103889092 | 0.13 |
C12orf42 |
chromosome 12 open reading frame 42 |
747 |
0.75 |
| chr8_145103879_145104096 | 0.13 |
CTD-3065J16.6 |
|
43 |
0.94 |
| chr16_32127388_32127983 | 0.13 |
IGHV3OR16-9 |
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
50299 |
0.11 |
| chr11_65360328_65360479 | 0.13 |
AP001362.1 |
Uncharacterized protein |
456 |
0.42 |
| chr8_82753589_82754405 | 0.13 |
SNX16 |
sorting nexin 16 |
196 |
0.96 |
| chr4_57521556_57521858 | 0.13 |
HOPX |
HOP homeobox |
763 |
0.73 |
| chr16_67281539_67281690 | 0.13 |
FHOD1 |
formin homology 2 domain containing 1 |
53 |
0.9 |
| chr1_110198687_110199384 | 0.13 |
GSTM4 |
glutathione S-transferase mu 4 |
77 |
0.94 |
| chr2_74709205_74709356 | 0.13 |
CCDC142 |
coiled-coil domain containing 142 |
843 |
0.25 |
| chr6_26087484_26088662 | 0.13 |
HFE |
hemochromatosis |
406 |
0.66 |
| chr18_657791_658473 | 0.13 |
C18orf56 |
chromosome 18 open reading frame 56 |
137 |
0.73 |
| chr15_90744552_90744900 | 0.13 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
42 |
0.96 |
| chr20_52789675_52790234 | 0.13 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
234 |
0.95 |
| chr7_100273526_100273689 | 0.13 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
137 |
0.92 |
| chr16_11038113_11038264 | 0.13 |
CLEC16A |
C-type lectin domain family 16, member A |
157 |
0.92 |
| chr21_47410185_47410336 | 0.13 |
COL6A1 |
collagen, type VI, alpha 1 |
8609 |
0.21 |
| chr12_96184496_96184659 | 0.13 |
NTN4 |
netrin 4 |
35 |
0.97 |
| chr9_139428442_139428593 | 0.13 |
RP11-413M3.4 |
|
8816 |
0.09 |
| chr11_279675_279826 | 0.13 |
NLRP6 |
NLR family, pyrin domain containing 6 |
1180 |
0.24 |
| chr5_132083512_132083663 | 0.13 |
CCNI2 |
cyclin I family, member 2 |
450 |
0.75 |
| chrX_25020829_25021251 | 0.13 |
ARX |
aristaless related homeobox |
13025 |
0.26 |
| chr3_52479164_52479315 | 0.13 |
SEMA3G |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
138 |
0.92 |
| chr17_7155597_7155825 | 0.13 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
16 |
0.54 |
| chr10_22623860_22624493 | 0.13 |
BMI1 |
BMI1 polycomb ring finger oncogene |
8728 |
0.15 |
| chr11_6341290_6341712 | 0.13 |
PRKCDBP |
protein kinase C, delta binding protein |
261 |
0.9 |
| chr8_23145924_23146455 | 0.13 |
R3HCC1 |
R3H domain and coiled-coil containing 1 |
254 |
0.9 |
| chr20_37229998_37230149 | 0.13 |
ARHGAP40 |
Rho GTPase activating protein 40 |
504 |
0.79 |
| chr11_6341817_6342265 | 0.13 |
PRKCDBP |
protein kinase C, delta binding protein |
164 |
0.94 |
| chr7_75988487_75988672 | 0.13 |
YWHAG |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
231 |
0.92 |
| chr15_96897228_96897556 | 0.13 |
AC087477.1 |
Uncharacterized protein |
7095 |
0.17 |
| chr19_20149762_20150000 | 0.13 |
ZNF682 |
zinc finger protein 682 |
133 |
0.96 |
| chr3_33759780_33760154 | 0.13 |
CLASP2 |
cytoplasmic linker associated protein 2 |
119 |
0.98 |
| chr10_64576967_64577230 | 0.13 |
EGR2 |
early growth response 2 |
972 |
0.57 |
| chr2_27309133_27309349 | 0.13 |
KHK |
ketohexokinase (fructokinase) |
374 |
0.67 |
| chr1_228400994_228401320 | 0.13 |
C1orf145 |
chromosome 1 open reading frame 145 |
204 |
0.9 |
| chr3_155588547_155589233 | 0.13 |
GMPS |
guanine monphosphate synthase |
451 |
0.85 |
| chr5_156692731_156693021 | 0.13 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
215 |
0.83 |
| chr7_102920201_102920819 | 0.13 |
DPY19L2P2 |
DPY19L2 pseudogene 2 |
2425 |
0.27 |
| chr18_24764849_24765000 | 0.13 |
CHST9 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
338 |
0.94 |
| chr8_65711600_65711819 | 0.13 |
CYP7B1 |
cytochrome P450, family 7, subfamily B, polypeptide 1 |
391 |
0.91 |
| chr6_11233064_11233578 | 0.13 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
406 |
0.89 |
| chr14_105144247_105144490 | 0.12 |
ENSG00000265291 |
. |
282 |
0.87 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 0.2 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.1 | 0.5 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.1 | 0.2 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.1 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.2 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.1 | 0.2 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.1 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.1 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.2 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.1 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0051797 | regulation of hair follicle development(GO:0051797) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0021612 | cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) |
| 0.0 | 0.1 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.2 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) |
| 0.0 | 0.0 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.2 | GO:0072665 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0060119 | inner ear receptor cell development(GO:0060119) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0060412 | ventricular septum morphogenesis(GO:0060412) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0048821 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.1 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0051307 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.2 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.3 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.1 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.1 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0072162 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.1 | GO:0043174 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) nucleoside salvage(GO:0043174) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:1903556 | negative regulation of tumor necrosis factor production(GO:0032720) negative regulation of tumor necrosis factor superfamily cytokine production(GO:1903556) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.1 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0051379 | epinephrine binding(GO:0051379) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.0 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 1.3 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.0 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.6 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |