Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
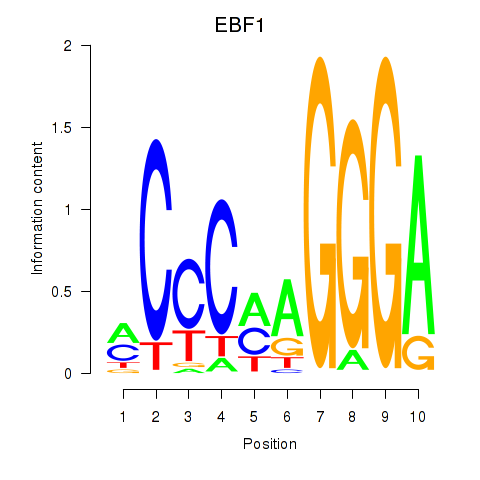
Results for EBF1
Z-value: 1.09
Transcription factors associated with EBF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF1
|
ENSG00000164330.12 | EBF transcription factor 1 |
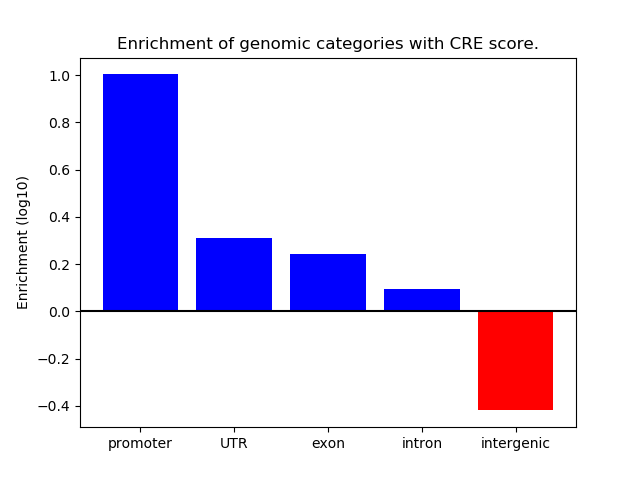
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_158532158_158532309 | EBF1 | 5464 | 0.273373 | -0.80 | 8.9e-03 | Click! |
| chr5_158518406_158518557 | EBF1 | 8220 | 0.259216 | -0.59 | 9.4e-02 | Click! |
| chr5_158479447_158479665 | EBF1 | 47145 | 0.146292 | 0.46 | 2.1e-01 | Click! |
| chr5_158533512_158533971 | EBF1 | 6972 | 0.261267 | 0.46 | 2.1e-01 | Click! |
| chr5_158478044_158478213 | EBF1 | 48573 | 0.142203 | 0.39 | 3.0e-01 | Click! |
Activity of the EBF1 motif across conditions
Conditions sorted by the z-value of the EBF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
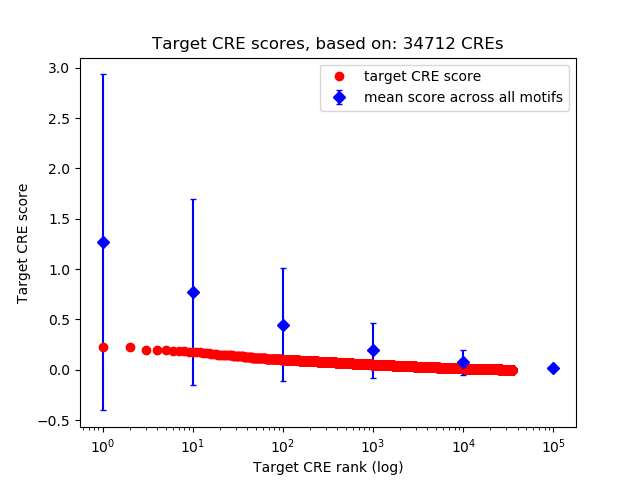
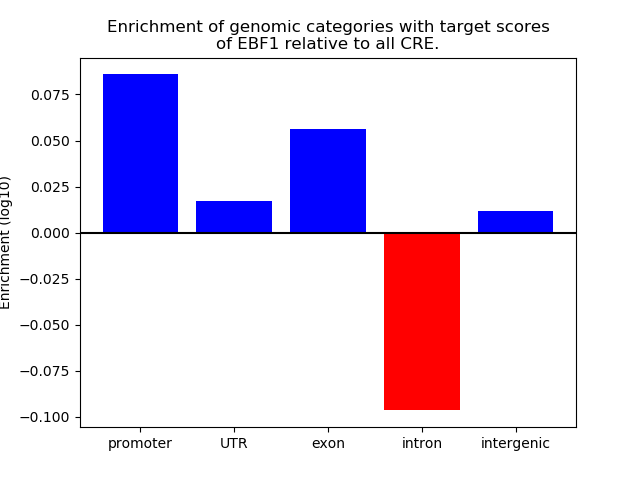
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_114136713_114136864 | 0.23 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
831 |
0.63 |
| chr17_61784423_61784574 | 0.23 |
STRADA |
STE20-related kinase adaptor alpha |
2683 |
0.21 |
| chr6_166954866_166955076 | 0.20 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
967 |
0.56 |
| chr2_97439727_97440064 | 0.20 |
CNNM4 |
cyclin M4 |
13256 |
0.14 |
| chr11_18728014_18728472 | 0.19 |
RP11-1081L13.4 |
|
159 |
0.94 |
| chr1_26644827_26645344 | 0.19 |
UBXN11 |
UBX domain protein 11 |
231 |
0.76 |
| chr4_7972052_7972961 | 0.18 |
AFAP1 |
actin filament associated protein 1 |
30853 |
0.14 |
| chr19_18112722_18113118 | 0.18 |
ARRDC2 |
arrestin domain containing 2 |
979 |
0.48 |
| chr15_89437756_89438448 | 0.18 |
HAPLN3 |
hyaluronan and proteoglycan link protein 3 |
662 |
0.7 |
| chr5_1105065_1105874 | 0.18 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
6681 |
0.19 |
| chr9_137417944_137418506 | 0.17 |
COL5A1 |
collagen, type V, alpha 1 |
115395 |
0.05 |
| chr19_10628197_10628587 | 0.17 |
S1PR5 |
sphingosine-1-phosphate receptor 5 |
215 |
0.87 |
| chr14_22957632_22957783 | 0.17 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
1536 |
0.24 |
| chr9_139776272_139777152 | 0.17 |
TRAF2 |
TNF receptor-associated factor 2 |
327 |
0.72 |
| chrX_130912849_130913434 | 0.16 |
ENSG00000200587 |
. |
159843 |
0.04 |
| chr2_10375130_10375281 | 0.16 |
ENSG00000265418 |
. |
42408 |
0.11 |
| chr22_19273287_19273568 | 0.16 |
CLTCL1 |
clathrin, heavy chain-like 1 |
5781 |
0.19 |
| chr2_236419917_236420068 | 0.15 |
AGAP1-IT1 |
AGAP1 intronic transcript 1 (non-protein coding) |
5597 |
0.24 |
| chr5_6828115_6828430 | 0.15 |
ENSG00000200243 |
. |
20303 |
0.27 |
| chr1_21619720_21620176 | 0.15 |
RP5-1071N3.1 |
|
165 |
0.91 |
| chrX_130875529_130875933 | 0.15 |
ENSG00000200587 |
. |
197253 |
0.03 |
| chr11_119191508_119192663 | 0.15 |
ENSG00000252119 |
. |
2034 |
0.16 |
| chr13_21807322_21807478 | 0.15 |
MRP63 |
mitochondrial ribosomal protein 63 |
56616 |
0.09 |
| chr5_38468727_38468878 | 0.14 |
CTD-2263F21.1 |
|
361 |
0.86 |
| chr15_56336229_56336380 | 0.14 |
ENSG00000239703 |
. |
14125 |
0.23 |
| chr8_99960098_99960383 | 0.14 |
OSR2 |
odd-skipped related transciption factor 2 |
210 |
0.93 |
| chrX_130917856_130918233 | 0.14 |
ENSG00000200587 |
. |
154940 |
0.04 |
| chr17_38483011_38483310 | 0.14 |
RARA |
retinoic acid receptor, alpha |
8623 |
0.11 |
| chr1_203713535_203713686 | 0.14 |
ENSG00000221643 |
. |
14901 |
0.15 |
| chr13_113555280_113555431 | 0.14 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
1152 |
0.51 |
| chr9_98994971_98995126 | 0.14 |
ENSG00000265367 |
. |
3156 |
0.24 |
| chr7_75911448_75912023 | 0.14 |
SRRM3 |
serine/arginine repetitive matrix 3 |
197 |
0.93 |
| chr17_74022610_74022808 | 0.14 |
EVPL |
envoplakin |
603 |
0.59 |
| chr22_50050359_50050996 | 0.14 |
C22orf34 |
chromosome 22 open reading frame 34 |
401 |
0.87 |
| chr20_43438673_43438824 | 0.13 |
RIMS4 |
regulating synaptic membrane exocytosis 4 |
164 |
0.95 |
| chr14_93531071_93531437 | 0.13 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
1449 |
0.4 |
| chr11_118212573_118212916 | 0.13 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
602 |
0.59 |
| chr1_201465199_201465350 | 0.13 |
CSRP1 |
cysteine and glycine-rich protein 1 |
427 |
0.81 |
| chr13_114906167_114906332 | 0.13 |
RASA3 |
RAS p21 protein activator 3 |
8163 |
0.22 |
| chr17_15174002_15174153 | 0.13 |
PMP22 |
peripheral myelin protein 22 |
5434 |
0.16 |
| chr19_36500441_36500617 | 0.13 |
SYNE4 |
spectrin repeat containing, nuclear envelope family member 4 |
834 |
0.4 |
| chr16_85769625_85769776 | 0.12 |
ENSG00000222190 |
. |
5606 |
0.13 |
| chr2_206546644_206547002 | 0.12 |
NRP2 |
neuropilin 2 |
401 |
0.91 |
| chr1_210517040_210517207 | 0.12 |
HHAT |
hedgehog acyltransferase |
14479 |
0.23 |
| chr5_139948767_139948946 | 0.12 |
SLC35A4 |
solute carrier family 35, member A4 |
4380 |
0.09 |
| chr1_32706125_32707286 | 0.12 |
MTMR9LP |
myotubularin related protein 9-like, pseudogene |
488 |
0.6 |
| chr19_41037225_41037376 | 0.12 |
SPTBN4 |
spectrin, beta, non-erythrocytic 4 |
905 |
0.52 |
| chr1_46982968_46983119 | 0.12 |
DMBX1 |
diencephalon/mesencephalon homeobox 1 |
10374 |
0.15 |
| chr20_44972040_44972274 | 0.12 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
14988 |
0.19 |
| chr20_1246931_1247537 | 0.12 |
SNPH |
syntaphilin |
235 |
0.91 |
| chr21_46351160_46352026 | 0.12 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
311 |
0.78 |
| chr1_151811855_151812405 | 0.12 |
C2CD4D |
C2 calcium-dependent domain containing 4D |
903 |
0.32 |
| chr19_35630816_35630967 | 0.12 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
35 |
0.95 |
| chr12_132689714_132689890 | 0.12 |
GALNT9 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) |
535 |
0.8 |
| chr14_52535921_52536493 | 0.12 |
NID2 |
nidogen 2 (osteonidogen) |
495 |
0.84 |
| chr1_226049341_226049492 | 0.12 |
TMEM63A |
transmembrane protein 63A |
5032 |
0.15 |
| chr11_118793309_118793854 | 0.12 |
BCL9L |
B-cell CLL/lymphoma 9-like |
3968 |
0.1 |
| chr1_88383945_88384194 | 0.12 |
ENSG00000199318 |
. |
465013 |
0.01 |
| chr12_117298021_117298350 | 0.11 |
HRK |
harakiri, BCL2 interacting protein (contains only BH3 domain) |
20708 |
0.2 |
| chr14_77393350_77393819 | 0.11 |
ENSG00000223174 |
. |
36978 |
0.14 |
| chr2_241949003_241950045 | 0.11 |
AC093585.6 |
|
637 |
0.62 |
| chr14_99725147_99725681 | 0.11 |
AL109767.1 |
|
3871 |
0.26 |
| chr1_154301309_154302009 | 0.11 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
384 |
0.73 |
| chr3_66479459_66479651 | 0.11 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
71801 |
0.12 |
| chr2_75904423_75904574 | 0.11 |
GCFC2 |
GC-rich sequence DNA-binding factor 2 |
7158 |
0.13 |
| chr1_85557311_85557759 | 0.11 |
WDR63 |
WD repeat domain 63 |
29525 |
0.15 |
| chr11_413234_413456 | 0.11 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
1603 |
0.21 |
| chr22_50908966_50909262 | 0.11 |
SBF1 |
SET binding factor 1 |
4257 |
0.1 |
| chr9_92083834_92084001 | 0.11 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
10888 |
0.23 |
| chr9_112888297_112888507 | 0.11 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
621 |
0.82 |
| chr1_161038546_161039545 | 0.11 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr17_41984276_41984918 | 0.11 |
MPP2 |
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
4 |
0.96 |
| chr2_98334586_98334804 | 0.11 |
ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
4672 |
0.2 |
| chr16_67282110_67283381 | 0.11 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
108 |
0.89 |
| chr2_236252173_236252324 | 0.11 |
ENSG00000216002 |
. |
17246 |
0.28 |
| chr17_9018882_9019405 | 0.11 |
NTN1 |
netrin 1 |
47109 |
0.15 |
| chr2_239036083_239036322 | 0.11 |
ESPNL |
espin-like |
166 |
0.94 |
| chr7_73413897_73414048 | 0.11 |
ELN |
elastin |
28147 |
0.17 |
| chr14_94437286_94437966 | 0.11 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
2803 |
0.22 |
| chr10_47640131_47640292 | 0.11 |
ANTXRLP1 |
anthrax toxin receptor-like pseudogene 1 |
4580 |
0.24 |
| chr14_73395581_73395732 | 0.11 |
DCAF4 |
DDB1 and CUL4 associated factor 4 |
2461 |
0.26 |
| chr2_112433070_112433221 | 0.11 |
ENSG00000266063 |
. |
95566 |
0.09 |
| chr22_33454563_33454714 | 0.11 |
SYN3 |
synapsin III |
280 |
0.95 |
| chr14_96593544_96593695 | 0.11 |
ENSG00000221236 |
. |
9569 |
0.19 |
| chr12_53600321_53600967 | 0.11 |
ITGB7 |
integrin, beta 7 |
356 |
0.78 |
| chr16_1047518_1048062 | 0.10 |
SOX8 |
SRY (sex determining region Y)-box 8 |
15982 |
0.12 |
| chr21_38352575_38353327 | 0.10 |
HLCS |
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
23 |
0.97 |
| chr17_4945612_4945862 | 0.10 |
SLC52A1 |
solute carrier family 52 (riboflavin transporter), member 1 |
7010 |
0.09 |
| chr3_58359199_58359350 | 0.10 |
PXK |
PX domain containing serine/threonine kinase |
22127 |
0.17 |
| chr5_134366245_134366951 | 0.10 |
PITX1 |
paired-like homeodomain 1 |
2227 |
0.27 |
| chr19_36004816_36005044 | 0.10 |
DMKN |
dermokine |
370 |
0.74 |
| chr17_75302303_75302454 | 0.10 |
SEPT9 |
septin 9 |
13219 |
0.22 |
| chr1_143466699_143467561 | 0.10 |
ENSG00000272302 |
. |
29721 |
0.2 |
| chr7_73867238_73867432 | 0.10 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
785 |
0.67 |
| chr14_53835257_53835408 | 0.10 |
RP11-547D23.1 |
|
215260 |
0.02 |
| chr2_237551540_237552008 | 0.10 |
ACKR3 |
atypical chemokine receptor 3 |
73490 |
0.12 |
| chr8_67522944_67523356 | 0.10 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
1992 |
0.31 |
| chr3_19030814_19030965 | 0.10 |
ENSG00000251761 |
. |
34348 |
0.24 |
| chr1_162646903_162647054 | 0.10 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
44718 |
0.14 |
| chr11_722189_722340 | 0.10 |
AP006621.9 |
|
4783 |
0.09 |
| chr8_125739093_125740062 | 0.10 |
MTSS1 |
metastasis suppressor 1 |
280 |
0.94 |
| chr8_22437078_22437527 | 0.10 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
362 |
0.75 |
| chr11_67417906_67418392 | 0.10 |
ACY3 |
aspartoacylase (aminocyclase) 3 |
19 |
0.96 |
| chr8_123101856_123102033 | 0.10 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
448268 |
0.01 |
| chr1_110075008_110075602 | 0.10 |
GPR61 |
G protein-coupled receptor 61 |
7189 |
0.1 |
| chrX_115004520_115004902 | 0.10 |
RP1-241P17.1 |
|
47368 |
0.14 |
| chr6_134211072_134211606 | 0.10 |
TCF21 |
transcription factor 21 |
866 |
0.5 |
| chr16_10746808_10746959 | 0.10 |
TEKT5 |
tektin 5 |
37093 |
0.13 |
| chr1_20397097_20397248 | 0.10 |
PLA2G5 |
phospholipase A2, group V |
471 |
0.81 |
| chr18_77163786_77163984 | 0.10 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
3493 |
0.3 |
| chr22_17517069_17517450 | 0.10 |
GAB4 |
GRB2-associated binding protein family, member 4 |
28147 |
0.14 |
| chr9_73018771_73018949 | 0.10 |
KLF9 |
Kruppel-like factor 9 |
10680 |
0.27 |
| chr2_7570809_7570960 | 0.10 |
ENSG00000221255 |
. |
146088 |
0.05 |
| chr9_136075040_136075613 | 0.10 |
OBP2B |
odorant binding protein 2B |
9302 |
0.14 |
| chr2_11884173_11884474 | 0.10 |
AC012456.4 |
|
1104 |
0.39 |
| chr16_89631829_89632776 | 0.10 |
RPL13 |
ribosomal protein L13 |
3524 |
0.13 |
| chr11_96073413_96073903 | 0.10 |
ENSG00000266192 |
. |
944 |
0.55 |
| chr5_82863395_82863716 | 0.10 |
VCAN-AS1 |
VCAN antisense RNA 1 |
5422 |
0.29 |
| chr18_12011713_12011864 | 0.10 |
RP11-820I16.3 |
|
16851 |
0.15 |
| chr4_6690869_6691175 | 0.10 |
AC093323.1 |
Uncharacterized protein |
3167 |
0.19 |
| chr8_22409217_22409843 | 0.10 |
SORBS3 |
sorbin and SH3 domain containing 3 |
322 |
0.81 |
| chr11_117859065_117859216 | 0.10 |
IL10RA |
interleukin 10 receptor, alpha |
2031 |
0.32 |
| chr6_147322190_147322415 | 0.10 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
85281 |
0.11 |
| chr12_96635413_96635600 | 0.10 |
RP11-394J1.2 |
|
17755 |
0.19 |
| chr12_112438793_112438944 | 0.10 |
TMEM116 |
transmembrane protein 116 |
4921 |
0.18 |
| chr1_16085392_16086186 | 0.10 |
FBLIM1 |
filamin binding LIM protein 1 |
497 |
0.68 |
| chr15_78557114_78557783 | 0.10 |
DNAJA4 |
DnaJ (Hsp40) homolog, subfamily A, member 4 |
532 |
0.55 |
| chr9_133321154_133321362 | 0.10 |
ASS1 |
argininosuccinate synthase 1 |
879 |
0.54 |
| chr19_55983710_55983861 | 0.10 |
ZNF628 |
zinc finger protein 628 |
3914 |
0.1 |
| chr4_184263832_184264346 | 0.10 |
ENSG00000238596 |
. |
13638 |
0.14 |
| chr19_7402613_7402882 | 0.10 |
CTB-133G6.1 |
|
11101 |
0.16 |
| chr6_147428076_147428227 | 0.10 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
12448 |
0.3 |
| chr17_17817656_17817807 | 0.10 |
TOM1L2 |
target of myb1-like 2 (chicken) |
42353 |
0.11 |
| chr5_67830491_67830731 | 0.10 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
242215 |
0.02 |
| chr7_2851005_2851785 | 0.10 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
3497 |
0.32 |
| chr2_189494316_189494467 | 0.10 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
59581 |
0.15 |
| chr16_10672917_10673068 | 0.09 |
RP11-27M24.2 |
|
952 |
0.47 |
| chr7_962027_962293 | 0.09 |
ADAP1 |
ArfGAP with dual PH domains 1 |
1634 |
0.31 |
| chr17_47419551_47419937 | 0.09 |
ENSG00000251964 |
. |
16481 |
0.12 |
| chr1_11713861_11714012 | 0.09 |
FBXO44 |
F-box protein 44 |
496 |
0.56 |
| chr1_17878359_17878510 | 0.09 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
12104 |
0.25 |
| chr17_73522650_73522801 | 0.09 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
855 |
0.44 |
| chr13_113295022_113295173 | 0.09 |
C13orf35 |
chromosome 13 open reading frame 35 |
6261 |
0.23 |
| chr12_54402958_54403134 | 0.09 |
HOXC8 |
homeobox C8 |
214 |
0.8 |
| chr1_156785806_156786457 | 0.09 |
SH2D2A |
SH2 domain containing 2A |
495 |
0.51 |
| chr7_132300022_132300173 | 0.09 |
PLXNA4 |
plexin A4 |
33350 |
0.18 |
| chr19_42057824_42058331 | 0.09 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
2191 |
0.27 |
| chr22_46453843_46454104 | 0.09 |
RP6-109B7.3 |
|
3178 |
0.13 |
| chr11_35281999_35282201 | 0.09 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
5192 |
0.19 |
| chr1_229335449_229335600 | 0.09 |
RP5-1061H20.5 |
|
27785 |
0.17 |
| chr14_22966536_22967037 | 0.09 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
10615 |
0.1 |
| chr11_62169770_62169944 | 0.09 |
SCGB1A1 |
secretoglobin, family 1A, member 1 (uteroglobin) |
2718 |
0.17 |
| chr6_36281244_36281395 | 0.09 |
C6orf222 |
chromosome 6 open reading frame 222 |
23343 |
0.15 |
| chr1_27324350_27324577 | 0.09 |
TRNP1 |
TMF1-regulated nuclear protein 1 |
3653 |
0.19 |
| chr8_37552103_37552726 | 0.09 |
ZNF703 |
zinc finger protein 703 |
855 |
0.5 |
| chr11_36002459_36002610 | 0.09 |
RP5-916O11.2 |
|
8556 |
0.2 |
| chr6_75918243_75918708 | 0.09 |
COL12A1 |
collagen, type XII, alpha 1 |
2708 |
0.31 |
| chr8_99008131_99008282 | 0.09 |
MATN2 |
matrilin 2 |
20566 |
0.15 |
| chr17_74489731_74490033 | 0.09 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
598 |
0.59 |
| chr20_17731005_17731156 | 0.09 |
BANF2 |
barrier to autointegration factor 2 |
50475 |
0.13 |
| chr14_103234628_103234779 | 0.09 |
TRAF3 |
TNF receptor-associated factor 3 |
9110 |
0.19 |
| chr14_104889660_104889811 | 0.09 |
ENSG00000222761 |
. |
22350 |
0.23 |
| chr13_24828231_24828382 | 0.09 |
SPATA13-AS1 |
SPATA13 antisense RNA 1 |
271 |
0.9 |
| chr14_74724486_74725063 | 0.09 |
VSX2 |
visual system homeobox 2 |
18599 |
0.14 |
| chr7_150441383_150441534 | 0.09 |
GIMAP5 |
GTPase, IMAP family member 5 |
7022 |
0.17 |
| chr18_13136915_13137798 | 0.09 |
RP11-794M8.1 |
|
78968 |
0.08 |
| chr10_17034557_17035249 | 0.09 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
8709 |
0.3 |
| chrX_130886755_130887324 | 0.09 |
ENSG00000200587 |
. |
185945 |
0.03 |
| chr14_99733165_99733625 | 0.09 |
AL109767.1 |
|
4110 |
0.24 |
| chr21_46352912_46353063 | 0.09 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
1083 |
0.32 |
| chr17_75453810_75454179 | 0.09 |
SEPT9 |
septin 9 |
1546 |
0.33 |
| chr3_194293899_194294050 | 0.09 |
TMEM44-AS1 |
TMEM44 antisense RNA 1 |
10766 |
0.15 |
| chr12_6901564_6902297 | 0.09 |
CD4 |
CD4 molecule |
3196 |
0.1 |
| chr4_103972156_103972307 | 0.09 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
6868 |
0.22 |
| chr8_28930678_28930829 | 0.09 |
CTD-2647L4.5 |
|
5957 |
0.15 |
| chr9_71736397_71737145 | 0.09 |
TJP2 |
tight junction protein 2 |
547 |
0.84 |
| chr2_112178560_112178711 | 0.09 |
ENSG00000266139 |
. |
99967 |
0.08 |
| chr2_149627712_149627970 | 0.09 |
KIF5C |
kinesin family member 5C |
4978 |
0.21 |
| chr10_71399492_71399643 | 0.09 |
C10orf35 |
chromosome 10 open reading frame 35 |
9267 |
0.21 |
| chr8_135469745_135470192 | 0.09 |
ZFAT |
zinc finger and AT hook domain containing |
52457 |
0.18 |
| chr10_79613508_79613659 | 0.09 |
AL391421.1 |
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
13050 |
0.18 |
| chr20_57721681_57721954 | 0.09 |
ZNF831 |
zinc finger protein 831 |
44258 |
0.15 |
| chr6_27218801_27219293 | 0.09 |
PRSS16 |
protease, serine, 16 (thymus) |
2332 |
0.31 |
| chr18_56530098_56530506 | 0.09 |
ZNF532 |
zinc finger protein 532 |
122 |
0.96 |
| chr9_96720776_96721007 | 0.09 |
BARX1 |
BARX homeobox 1 |
3237 |
0.33 |
| chr14_22984887_22985075 | 0.09 |
TRAJ15 |
T cell receptor alpha joining 15 |
13599 |
0.1 |
| chr1_207996298_207996642 | 0.09 |
ENSG00000203709 |
. |
20602 |
0.22 |
| chr1_227070133_227070284 | 0.09 |
PSEN2 |
presenilin 2 (Alzheimer disease 4) |
1861 |
0.39 |
| chr3_38500973_38501124 | 0.09 |
ACVR2B-AS1 |
ACVR2B antisense RNA 1 |
4737 |
0.19 |
| chr13_44730254_44730535 | 0.09 |
SMIM2-IT1 |
SMIM2 intronic transcript 1 (non-protein coding) |
1964 |
0.33 |
| chr1_12677001_12677791 | 0.09 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
341 |
0.87 |
| chr5_156886857_156887334 | 0.09 |
CTB-109A12.1 |
|
9 |
0.54 |
| chr3_151922835_151923340 | 0.09 |
MBNL1 |
muscleblind-like splicing regulator 1 |
62742 |
0.13 |
| chr17_43299697_43300109 | 0.09 |
CTD-2020K17.1 |
|
314 |
0.65 |
| chr2_85107579_85108198 | 0.09 |
TRABD2A |
TraB domain containing 2A |
318 |
0.89 |
| chr10_86300062_86300319 | 0.09 |
CCSER2 |
coiled-coil serine-rich protein 2 |
115495 |
0.07 |
| chr9_126807806_126807963 | 0.09 |
RP11-85O21.5 |
|
13081 |
0.17 |
| chrX_1776580_1777384 | 0.09 |
ASMT |
acetylserotonin O-methyltransferase |
21691 |
0.23 |
| chr10_99932028_99932445 | 0.09 |
R3HCC1L |
R3H domain and coiled-coil containing 1-like |
9596 |
0.25 |
| chr5_138441478_138441629 | 0.09 |
SIL1 |
SIL1 nucleotide exchange factor |
25894 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0072539 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0044266 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0072162 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0070828 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.0 | GO:0045683 | negative regulation of epidermis development(GO:0045683) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.1 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:0002874 | chronic inflammatory response to antigenic stimulus(GO:0002439) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.0 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |