Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
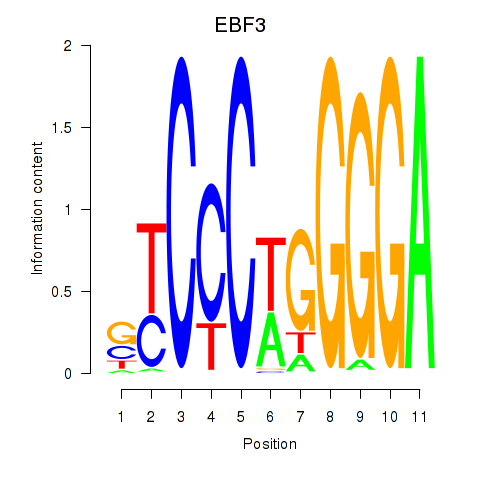
Results for EBF3
Z-value: 1.56
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.9 | EBF transcription factor 3 |
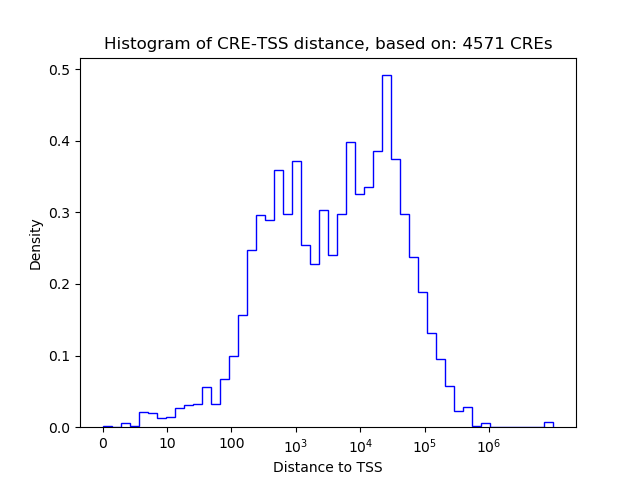
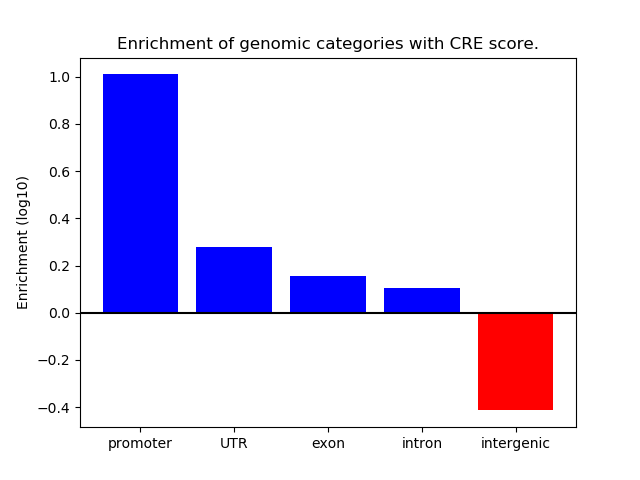
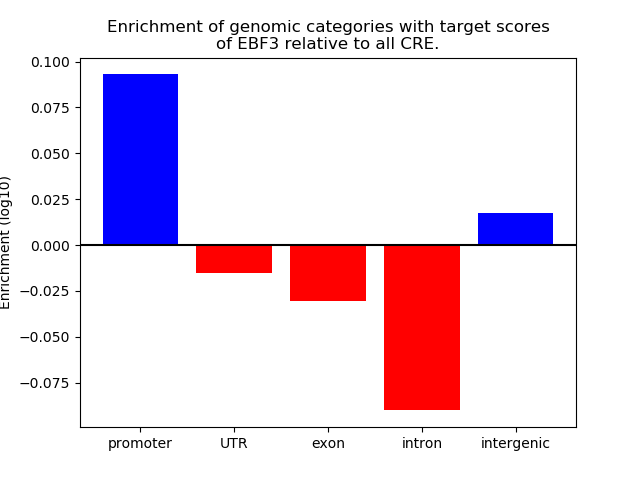
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_131751845_131752016 | EBF3 | 10175 | 0.301822 | 0.94 | 1.4e-04 | Click! |
| chr10_131744784_131744954 | EBF3 | 17236 | 0.279829 | 0.90 | 9.3e-04 | Click! |
| chr10_131759230_131759381 | EBF3 | 2800 | 0.404463 | 0.89 | 1.2e-03 | Click! |
| chr10_131759565_131759716 | EBF3 | 2465 | 0.428919 | 0.88 | 1.7e-03 | Click! |
| chr10_131811384_131811535 | EBF3 | 49354 | 0.181687 | 0.80 | 9.7e-03 | Click! |
Activity of the EBF3 motif across conditions
Conditions sorted by the z-value of the EBF3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
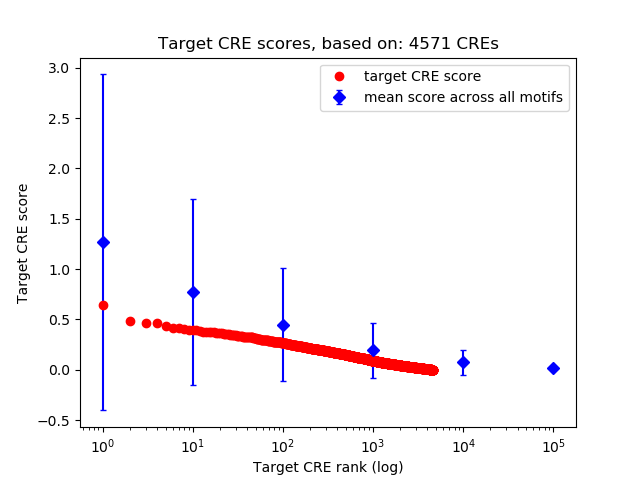
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_227661377_227661773 | 0.64 |
IRS1 |
insulin receptor substrate 1 |
2900 |
0.28 |
| chr19_50031610_50031873 | 0.48 |
RCN3 |
reticulocalbin 3, EF-hand calcium binding domain |
142 |
0.87 |
| chr2_177001412_177001990 | 0.47 |
HOXD3 |
homeobox D3 |
16 |
0.5 |
| chr3_100711576_100712203 | 0.46 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
408 |
0.89 |
| chr16_86020719_86020879 | 0.44 |
IRF8 |
interferon regulatory factor 8 |
72880 |
0.11 |
| chr17_29887299_29887674 | 0.42 |
ENSG00000207614 |
. |
471 |
0.69 |
| chr18_77225876_77226142 | 0.41 |
AC018445.1 |
Uncharacterized protein |
50048 |
0.15 |
| chr5_95768411_95768678 | 0.41 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
440 |
0.89 |
| chrX_17394576_17394820 | 0.40 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
1155 |
0.54 |
| chr3_61548960_61549133 | 0.39 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
1461 |
0.59 |
| chr2_176987899_176988234 | 0.39 |
HOXD9 |
homeobox D9 |
978 |
0.29 |
| chr9_129883603_129883754 | 0.38 |
ANGPTL2 |
angiopoietin-like 2 |
1235 |
0.56 |
| chr4_81195995_81196185 | 0.38 |
FGF5 |
fibroblast growth factor 5 |
8297 |
0.25 |
| chr1_168698177_168698379 | 0.38 |
DPT |
dermatopontin |
224 |
0.95 |
| chr2_230035602_230035855 | 0.37 |
PID1 |
phosphotyrosine interaction domain containing 1 |
61073 |
0.14 |
| chr14_69419495_69419882 | 0.37 |
ACTN1 |
actinin, alpha 1 |
5423 |
0.25 |
| chr20_56749874_56750278 | 0.37 |
C20orf85 |
chromosome 20 open reading frame 85 |
24116 |
0.19 |
| chr16_88471986_88472161 | 0.37 |
ZNF469 |
zinc finger protein 469 |
21806 |
0.18 |
| chr7_73183095_73183263 | 0.37 |
CLDN3 |
claudin 3 |
1421 |
0.3 |
| chr20_39969728_39969901 | 0.37 |
LPIN3 |
lipin 3 |
254 |
0.92 |
| chr9_126803162_126803381 | 0.36 |
RP11-85O21.5 |
|
8468 |
0.18 |
| chr8_49831422_49831573 | 0.36 |
SNAI2 |
snail family zinc finger 2 |
2491 |
0.43 |
| chr3_170139818_170139969 | 0.36 |
CLDN11 |
claudin 11 |
3038 |
0.33 |
| chr12_110270768_110271158 | 0.35 |
ENSG00000263510 |
. |
190 |
0.56 |
| chr11_132813237_132813595 | 0.35 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
150 |
0.98 |
| chr17_41613761_41613953 | 0.35 |
ETV4 |
ets variant 4 |
6055 |
0.15 |
| chr18_21851258_21851803 | 0.35 |
OSBPL1A |
oxysterol binding protein-like 1A |
666 |
0.6 |
| chr12_96183320_96183471 | 0.34 |
NTN4 |
netrin 4 |
331 |
0.86 |
| chr2_177000583_177000734 | 0.34 |
HOXD3 |
homeobox D3 |
1027 |
0.23 |
| chr2_3465758_3466116 | 0.34 |
TRAPPC12 |
trafficking protein particle complex 12 |
3507 |
0.21 |
| chr19_49219826_49219992 | 0.34 |
MAMSTR |
MEF2 activating motif and SAP domain containing transcriptional regulator |
191 |
0.87 |
| chr18_59401096_59401293 | 0.33 |
RNF152 |
ring finger protein 152 |
159798 |
0.04 |
| chr1_41962190_41962352 | 0.33 |
EDN2 |
endothelin 2 |
11929 |
0.2 |
| chr9_137535529_137535696 | 0.33 |
COL5A1 |
collagen, type V, alpha 1 |
1992 |
0.35 |
| chr16_4422135_4423224 | 0.33 |
VASN |
vasorin |
830 |
0.49 |
| chr1_203456320_203456508 | 0.33 |
OPTC |
opticin |
6857 |
0.22 |
| chr13_24477780_24477964 | 0.33 |
C1QTNF9B |
C1q and tumor necrosis factor related protein 9B |
1078 |
0.48 |
| chr9_14924390_14924574 | 0.33 |
FREM1 |
FRAS1 related extracellular matrix 1 |
13489 |
0.26 |
| chr7_101579659_101580126 | 0.33 |
CTB-181H17.1 |
|
23504 |
0.22 |
| chr12_54395536_54395687 | 0.33 |
HOXC9 |
homeobox C9 |
1708 |
0.12 |
| chr13_113555280_113555431 | 0.33 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
1152 |
0.51 |
| chr7_143208334_143208676 | 0.32 |
TAS2R41 |
taste receptor, type 2, member 41 |
33539 |
0.13 |
| chr10_86300062_86300319 | 0.32 |
CCSER2 |
coiled-coil serine-rich protein 2 |
115495 |
0.07 |
| chr14_20903201_20903901 | 0.32 |
KLHL33 |
kelch-like family member 33 |
250 |
0.81 |
| chr11_46307086_46307288 | 0.32 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
7959 |
0.18 |
| chr10_27914063_27914342 | 0.32 |
MKX |
mohawk homeobox |
118272 |
0.05 |
| chr8_6539884_6540241 | 0.32 |
CTD-2541M15.1 |
|
25638 |
0.18 |
| chr19_34973467_34973843 | 0.32 |
WTIP |
Wilms tumor 1 interacting protein |
222 |
0.92 |
| chr15_42749089_42749416 | 0.31 |
ZNF106 |
zinc finger protein 106 |
459 |
0.78 |
| chr7_157071827_157072074 | 0.31 |
ENSG00000266453 |
. |
26537 |
0.21 |
| chr1_40598519_40598964 | 0.31 |
RLF |
rearranged L-myc fusion |
28304 |
0.14 |
| chr7_155049844_155050012 | 0.31 |
INSIG1 |
insulin induced gene 1 |
39558 |
0.15 |
| chr17_70113469_70114161 | 0.31 |
SOX9 |
SRY (sex determining region Y)-box 9 |
3346 |
0.37 |
| chr1_201619846_201619997 | 0.30 |
NAV1 |
neuron navigator 1 |
2471 |
0.24 |
| chr19_36500441_36500617 | 0.30 |
SYNE4 |
spectrin repeat containing, nuclear envelope family member 4 |
834 |
0.4 |
| chr18_47172272_47172498 | 0.30 |
LIPG |
lipase, endothelial |
83670 |
0.07 |
| chr19_13618302_13618480 | 0.30 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
1074 |
0.61 |
| chr3_126398343_126398521 | 0.30 |
TXNRD3 |
thioredoxin reductase 3 |
24434 |
0.17 |
| chr19_49661333_49661989 | 0.30 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
562 |
0.58 |
| chr10_69867536_69867687 | 0.30 |
MYPN |
myopalladin |
1383 |
0.44 |
| chr22_46453843_46454104 | 0.30 |
RP6-109B7.3 |
|
3178 |
0.13 |
| chr4_111397812_111398104 | 0.30 |
ENPEP |
glutamyl aminopeptidase (aminopeptidase A) |
729 |
0.72 |
| chr3_126006139_126006425 | 0.30 |
KLF15 |
Kruppel-like factor 15 |
70003 |
0.08 |
| chr9_116327712_116328152 | 0.29 |
RGS3 |
regulator of G-protein signaling 3 |
571 |
0.8 |
| chr11_61657871_61658132 | 0.29 |
FADS3 |
fatty acid desaturase 3 |
852 |
0.49 |
| chr15_96903147_96903373 | 0.29 |
AC087477.1 |
Uncharacterized protein |
1227 |
0.45 |
| chr20_5484666_5484886 | 0.29 |
RP5-1022P6.5 |
|
29498 |
0.21 |
| chr7_130070545_130070874 | 0.29 |
CEP41 |
centrosomal protein 41kDa |
3889 |
0.17 |
| chr6_136360177_136360429 | 0.29 |
RP13-143G15.3 |
|
25473 |
0.21 |
| chr9_102828891_102829115 | 0.29 |
ERP44 |
endoplasmic reticulum protein 44 |
32319 |
0.16 |
| chr19_46916092_46916684 | 0.29 |
CCDC8 |
coiled-coil domain containing 8 |
453 |
0.75 |
| chr5_82863395_82863716 | 0.29 |
VCAN-AS1 |
VCAN antisense RNA 1 |
5422 |
0.29 |
| chr1_1355307_1355467 | 0.29 |
RP4-758J18.7 |
|
238 |
0.74 |
| chr9_137534558_137534843 | 0.28 |
COL5A1 |
collagen, type V, alpha 1 |
1080 |
0.55 |
| chr19_19050618_19051263 | 0.28 |
HOMER3 |
homer homolog 3 (Drosophila) |
173 |
0.5 |
| chr16_67634950_67635107 | 0.28 |
AC009095.4 |
|
14217 |
0.09 |
| chr15_71148348_71148634 | 0.28 |
LARP6 |
La ribonucleoprotein domain family, member 6 |
1993 |
0.26 |
| chr8_19459992_19460252 | 0.28 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
116 |
0.98 |
| chr2_9879900_9880051 | 0.28 |
ENSG00000200034 |
. |
937 |
0.66 |
| chr11_27720868_27721098 | 0.28 |
BDNF |
brain-derived neurotrophic factor |
231 |
0.95 |
| chr6_3517843_3518042 | 0.28 |
SLC22A23 |
solute carrier family 22, member 23 |
60686 |
0.14 |
| chr2_145278908_145279072 | 0.28 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
369 |
0.75 |
| chr17_75318226_75318377 | 0.28 |
SEPT9 |
septin 9 |
391 |
0.88 |
| chr3_64670816_64671530 | 0.28 |
ADAMTS9-AS2 |
ADAMTS9 antisense RNA 2 |
369 |
0.87 |
| chr1_23079970_23080247 | 0.28 |
ENSG00000216157 |
. |
4704 |
0.19 |
| chr14_24803093_24804202 | 0.28 |
ADCY4 |
adenylate cyclase 4 |
306 |
0.72 |
| chrX_17393577_17393934 | 0.28 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
212 |
0.95 |
| chr9_89558891_89559042 | 0.27 |
GAS1 |
growth arrest-specific 1 |
3138 |
0.4 |
| chr17_65486818_65487236 | 0.27 |
ENSG00000207688 |
. |
19422 |
0.14 |
| chr2_172448669_172448823 | 0.27 |
CYBRD1 |
cytochrome b reductase 1 |
69157 |
0.11 |
| chr4_86398617_86398768 | 0.27 |
ARHGAP24 |
Rho GTPase activating protein 24 |
2301 |
0.46 |
| chr1_226001327_226001516 | 0.27 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
3585 |
0.18 |
| chr3_38071745_38071896 | 0.27 |
PLCD1 |
phospholipase C, delta 1 |
567 |
0.68 |
| chr17_76330950_76331176 | 0.27 |
SOCS3 |
suppressor of cytokine signaling 3 |
25092 |
0.13 |
| chr14_25518126_25518277 | 0.27 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
5 |
0.99 |
| chr19_4575354_4575624 | 0.27 |
SEMA6B |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
15669 |
0.1 |
| chr17_56494859_56495034 | 0.27 |
RNF43 |
ring finger protein 43 |
3 |
0.97 |
| chr17_12888079_12888251 | 0.27 |
ARHGAP44 |
Rho GTPase activating protein 44 |
4740 |
0.18 |
| chr1_31380843_31381078 | 0.27 |
SDC3 |
syndecan 3 |
648 |
0.66 |
| chr2_200320371_200320723 | 0.27 |
SATB2 |
SATB homeobox 2 |
264 |
0.92 |
| chr17_25821548_25821771 | 0.27 |
KSR1 |
kinase suppressor of ras 1 |
22623 |
0.18 |
| chr8_26308983_26309354 | 0.27 |
RP11-14I17.3 |
|
11161 |
0.19 |
| chr2_151342879_151343387 | 0.27 |
RND3 |
Rho family GTPase 3 |
1048 |
0.71 |
| chr20_17875766_17876211 | 0.26 |
ENSG00000212555 |
. |
8418 |
0.2 |
| chr14_103522624_103522803 | 0.26 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
1086 |
0.51 |
| chr19_47921712_47921897 | 0.26 |
MEIS3 |
Meis homeobox 3 |
575 |
0.72 |
| chr11_19617803_19618188 | 0.26 |
ENSG00000207407 |
. |
5157 |
0.23 |
| chrX_53349214_53349365 | 0.26 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
1233 |
0.43 |
| chr20_56622065_56622292 | 0.26 |
ENSG00000221385 |
. |
20334 |
0.22 |
| chr19_46270378_46270670 | 0.26 |
AC074212.6 |
|
530 |
0.46 |
| chr16_46663412_46663563 | 0.26 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
7949 |
0.18 |
| chr2_128439019_128439205 | 0.26 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
248 |
0.91 |
| chr19_16436739_16437476 | 0.26 |
KLF2 |
Kruppel-like factor 2 |
1456 |
0.3 |
| chr16_53451316_53451467 | 0.26 |
RBL2 |
retinoblastoma-like 2 (p130) |
16498 |
0.15 |
| chr18_9594992_9595154 | 0.26 |
RP11-881L2.1 |
|
7689 |
0.17 |
| chr12_49741682_49741986 | 0.25 |
DNAJC22 |
DnaJ (Hsp40) homolog, subfamily C, member 22 |
1013 |
0.39 |
| chr16_65154751_65155029 | 0.25 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
943 |
0.75 |
| chr8_97506014_97506898 | 0.25 |
SDC2 |
syndecan 2 |
220 |
0.96 |
| chr3_73673186_73673337 | 0.25 |
PDZRN3-AS1 |
PDZRN3 antisense RNA 1 |
542 |
0.56 |
| chr10_118500836_118501537 | 0.25 |
HSPA12A |
heat shock 70kDa protein 12A |
899 |
0.64 |
| chr6_39614678_39614962 | 0.25 |
KIF6 |
kinesin family member 6 |
7361 |
0.26 |
| chr17_38500769_38502147 | 0.25 |
RARA |
retinoic acid receptor, alpha |
35 |
0.95 |
| chr19_4058568_4058732 | 0.25 |
CTD-2622I13.3 |
|
4097 |
0.13 |
| chr19_3404785_3405081 | 0.25 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
38349 |
0.11 |
| chr14_87325066_87325217 | 0.25 |
RP11-322L20.1 |
HCG2028865; Uncharacterized protein |
46981 |
0.21 |
| chr2_9953892_9954060 | 0.25 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
29507 |
0.19 |
| chr2_223919112_223919263 | 0.25 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
2325 |
0.41 |
| chr1_201436884_201437966 | 0.25 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
887 |
0.55 |
| chr6_127440959_127441265 | 0.24 |
RSPO3 |
R-spondin 3 |
929 |
0.72 |
| chr9_88861790_88861958 | 0.24 |
ENSG00000223012 |
. |
11687 |
0.19 |
| chr3_65342341_65342492 | 0.24 |
RP11-88H12.2 |
|
2976 |
0.39 |
| chr7_47367963_47368180 | 0.24 |
TNS3 |
tensin 3 |
48155 |
0.2 |
| chr10_74878153_74878313 | 0.24 |
RP11-152N13.16 |
|
6930 |
0.1 |
| chr19_14606640_14606859 | 0.24 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
195 |
0.89 |
| chr17_15016441_15016644 | 0.24 |
ENSG00000238806 |
. |
124658 |
0.05 |
| chr5_134366245_134366951 | 0.24 |
PITX1 |
paired-like homeodomain 1 |
2227 |
0.27 |
| chr19_524423_524574 | 0.24 |
CDC34 |
cell division cycle 34 |
7216 |
0.09 |
| chr7_99835180_99835357 | 0.24 |
ENSG00000222482 |
. |
17525 |
0.08 |
| chr1_88383945_88384194 | 0.24 |
ENSG00000199318 |
. |
465013 |
0.01 |
| chr22_24383812_24383984 | 0.24 |
GSTT1 |
glutathione S-transferase theta 1 |
344 |
0.8 |
| chr1_149908154_149908323 | 0.24 |
MTMR11 |
myotubularin related protein 11 |
5 |
0.95 |
| chr10_102278895_102279852 | 0.24 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
218 |
0.92 |
| chr19_47524215_47524432 | 0.24 |
NPAS1 |
neuronal PAS domain protein 1 |
180 |
0.94 |
| chr19_46117807_46118118 | 0.24 |
EML2 |
echinoderm microtubule associated protein like 2 |
4386 |
0.11 |
| chr7_97839825_97840267 | 0.24 |
BHLHA15 |
basic helix-loop-helix family, member a15 |
693 |
0.72 |
| chr18_60845880_60846088 | 0.24 |
ENSG00000238988 |
. |
15914 |
0.21 |
| chr1_227635446_227635597 | 0.24 |
ENSG00000202264 |
. |
113361 |
0.06 |
| chr19_55795582_55795793 | 0.24 |
BRSK1 |
BR serine/threonine kinase 1 |
102 |
0.92 |
| chr6_5498160_5498518 | 0.24 |
RP1-232P20.1 |
|
40031 |
0.2 |
| chr14_69015576_69015827 | 0.24 |
CTD-2325P2.4 |
|
79461 |
0.1 |
| chr12_121345014_121345220 | 0.24 |
SPPL3 |
signal peptide peptidase like 3 |
2943 |
0.25 |
| chr22_33698264_33698426 | 0.24 |
SC22CB-1D7.1 |
|
18506 |
0.25 |
| chr10_93472068_93472249 | 0.23 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
79347 |
0.09 |
| chr14_69950443_69950594 | 0.23 |
PLEKHD1 |
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
953 |
0.65 |
| chr17_75319506_75319975 | 0.23 |
SEPT9 |
septin 9 |
1048 |
0.58 |
| chr1_156659032_156659371 | 0.23 |
NES |
nestin |
12012 |
0.08 |
| chr10_84741142_84741483 | 0.23 |
ENSG00000200774 |
. |
86515 |
0.11 |
| chr6_1575620_1576144 | 0.23 |
FOXC1 |
forkhead box C1 |
34799 |
0.21 |
| chr10_92680065_92680221 | 0.23 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
890 |
0.55 |
| chr3_184301056_184301262 | 0.23 |
EPHB3 |
EPH receptor B3 |
21587 |
0.18 |
| chr15_77788917_77789110 | 0.23 |
HMG20A |
high mobility group 20A |
18378 |
0.24 |
| chr5_142592854_142593005 | 0.23 |
ARHGAP26 |
Rho GTPase activating protein 26 |
6164 |
0.27 |
| chr3_134050593_134051228 | 0.23 |
AMOTL2 |
angiomotin like 2 |
39844 |
0.16 |
| chrX_67867525_67867904 | 0.23 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
104 |
0.98 |
| chr7_18810538_18810689 | 0.23 |
ENSG00000222164 |
. |
37289 |
0.22 |
| chr3_184893391_184893626 | 0.23 |
EHHADH-AS1 |
EHHADH antisense RNA 1 |
12819 |
0.2 |
| chr12_127867879_127868128 | 0.23 |
ENSG00000253089 |
. |
62103 |
0.16 |
| chr4_41015538_41015689 | 0.23 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
789 |
0.67 |
| chr5_71453247_71453481 | 0.23 |
ENSG00000264099 |
. |
11930 |
0.23 |
| chr5_64438498_64438671 | 0.23 |
ENSG00000207439 |
. |
19388 |
0.29 |
| chr15_45933944_45934448 | 0.23 |
SQRDL |
sulfide quinone reductase-like (yeast) |
3883 |
0.22 |
| chr15_81072135_81072315 | 0.23 |
KIAA1199 |
KIAA1199 |
513 |
0.83 |
| chr1_9257961_9258177 | 0.23 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
36765 |
0.13 |
| chr2_239036083_239036322 | 0.23 |
ESPNL |
espin-like |
166 |
0.94 |
| chr2_238321225_238321570 | 0.23 |
COL6A3 |
collagen, type VI, alpha 3 |
1394 |
0.45 |
| chrX_139846676_139846867 | 0.23 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
19952 |
0.22 |
| chr20_45583383_45583578 | 0.23 |
EYA2 |
eyes absent homolog 2 (Drosophila) |
35160 |
0.2 |
| chr1_211306419_211307099 | 0.23 |
KCNH1-IT1 |
KCNH1 intronic transcript 1 (non-protein coding) |
43 |
0.92 |
| chr11_8740394_8740932 | 0.22 |
ST5 |
suppression of tumorigenicity 5 |
27 |
0.96 |
| chr5_138289219_138289395 | 0.22 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
22980 |
0.17 |
| chr7_1028148_1028407 | 0.22 |
CYP2W1 |
cytochrome P450, family 2, subfamily W, polypeptide 1 |
1984 |
0.21 |
| chr12_54430598_54430749 | 0.22 |
ENSG00000207571 |
. |
2939 |
0.1 |
| chr15_23454167_23454360 | 0.22 |
ENSG00000238347 |
. |
9495 |
0.15 |
| chr11_66102741_66104078 | 0.22 |
RIN1 |
Ras and Rab interactor 1 |
468 |
0.53 |
| chr1_210517040_210517207 | 0.22 |
HHAT |
hedgehog acyltransferase |
14479 |
0.23 |
| chr11_8739823_8739992 | 0.22 |
ST5 |
suppression of tumorigenicity 5 |
117 |
0.94 |
| chr11_89867468_89867846 | 0.22 |
NAALAD2 |
N-acetylated alpha-linked acidic dipeptidase 2 |
40 |
0.98 |
| chr17_78194664_78195096 | 0.22 |
SGSH |
N-sulfoglucosamine sulfohydrolase |
158 |
0.63 |
| chr12_106533400_106533896 | 0.22 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
163 |
0.97 |
| chr10_52750817_52751095 | 0.22 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
11 |
0.99 |
| chr9_16867910_16868386 | 0.22 |
BNC2 |
basonuclin 2 |
2556 |
0.42 |
| chr16_2210764_2211128 | 0.22 |
TRAF7 |
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
2597 |
0.09 |
| chr10_119294315_119294466 | 0.22 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
7440 |
0.24 |
| chr4_121991394_121991545 | 0.22 |
NDNF |
neuron-derived neurotrophic factor |
1687 |
0.41 |
| chr16_54970178_54971110 | 0.22 |
IRX5 |
iroquois homeobox 5 |
4660 |
0.36 |
| chr17_1903139_1903290 | 0.22 |
CTD-2545H1.2 |
|
4306 |
0.12 |
| chr16_80972630_80972781 | 0.22 |
CMC2 |
C-x(9)-C motif containing 2 |
59559 |
0.09 |
| chr8_21990263_21990450 | 0.22 |
HR |
hair growth associated |
975 |
0.39 |
| chr10_90710785_90711179 | 0.22 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
1548 |
0.33 |
| chr2_120189004_120189226 | 0.22 |
TMEM37 |
transmembrane protein 37 |
330 |
0.89 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.4 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.3 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.1 | 0.3 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.1 | 0.3 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.2 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.4 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.2 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.1 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.6 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0060602 | branch elongation of an epithelium(GO:0060602) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.0 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.3 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:1990845 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0060444 | branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0060603 | mammary gland duct morphogenesis(GO:0060603) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.3 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.0 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:1904748 | regulation of apoptotic process involved in morphogenesis(GO:1902337) regulation of apoptotic process involved in development(GO:1904748) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.0 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0060536 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0072538 | T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.2 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.4 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0052509 | modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.0 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.0 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0044217 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.3 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.1 | 0.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.4 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.2 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |