Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
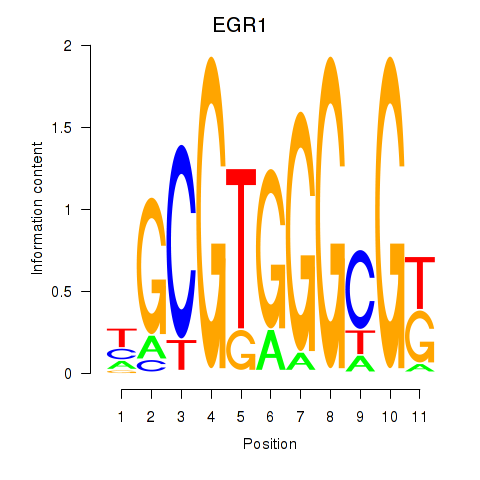
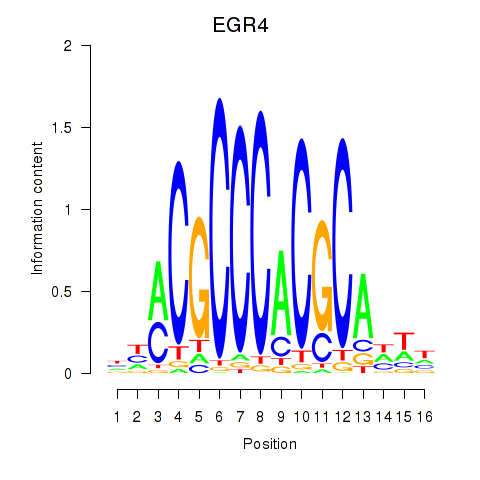
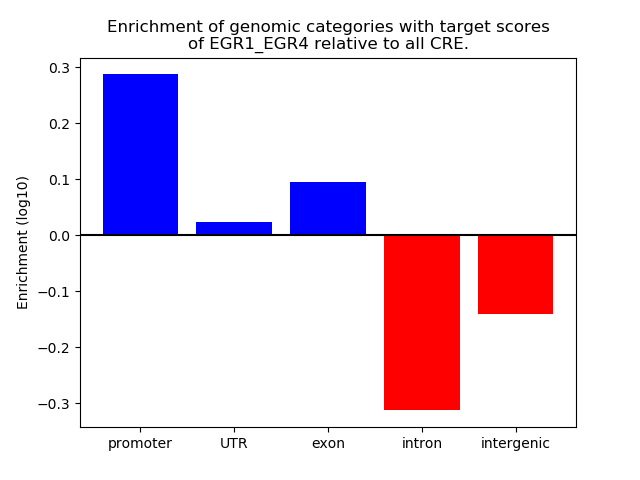
Results for EGR1_EGR4
Z-value: 0.95
Transcription factors associated with EGR1_EGR4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR1
|
ENSG00000120738.7 | early growth response 1 |
|
EGR4
|
ENSG00000135625.6 | early growth response 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_137802391_137802557 | EGR1 | 1295 | 0.412483 | 0.75 | 1.9e-02 | Click! |
| chr5_137802209_137802360 | EGR1 | 1105 | 0.470478 | 0.73 | 2.6e-02 | Click! |
| chr5_137805025_137805176 | EGR1 | 3921 | 0.199444 | 0.67 | 4.8e-02 | Click! |
| chr5_137803301_137803461 | EGR1 | 2202 | 0.270567 | 0.62 | 7.5e-02 | Click! |
| chr5_137801989_137802208 | EGR1 | 919 | 0.543403 | 0.59 | 9.6e-02 | Click! |
| chr2_73518392_73518872 | EGR4 | 2197 | 0.234160 | 0.54 | 1.4e-01 | Click! |
| chr2_73520179_73521065 | EGR4 | 207 | 0.917132 | 0.47 | 2.1e-01 | Click! |
| chr2_73521076_73521227 | EGR4 | 318 | 0.855192 | 0.31 | 4.1e-01 | Click! |
| chr2_73519153_73519304 | EGR4 | 1601 | 0.303270 | -0.18 | 6.5e-01 | Click! |
Activity of the EGR1_EGR4 motif across conditions
Conditions sorted by the z-value of the EGR1_EGR4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_133308330_133308718 | 0.57 |
HMCN2 |
hemicentin 2 |
2855 |
0.21 |
| chr14_75077508_75077674 | 0.54 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
1184 |
0.5 |
| chr3_55514769_55515136 | 0.45 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
272 |
0.94 |
| chr9_124480535_124480686 | 0.45 |
DAB2IP |
DAB2 interacting protein |
18961 |
0.24 |
| chr17_62051691_62051850 | 0.41 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
1492 |
0.3 |
| chr22_37730993_37731243 | 0.40 |
CYTH4 |
cytohesin 4 |
25109 |
0.13 |
| chr15_96897228_96897556 | 0.39 |
AC087477.1 |
Uncharacterized protein |
7095 |
0.17 |
| chr11_69191812_69191963 | 0.36 |
MYEOV |
myeloma overexpressed |
130262 |
0.05 |
| chr13_20735020_20735550 | 0.35 |
GJA3 |
gap junction protein, alpha 3, 46kDa |
97 |
0.97 |
| chr1_236305972_236306453 | 0.34 |
GPR137B |
G protein-coupled receptor 137B |
144 |
0.97 |
| chr4_99214803_99214954 | 0.32 |
RP11-323J4.1 |
|
32097 |
0.19 |
| chrX_110339181_110339857 | 0.30 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
69 |
0.99 |
| chr21_28339057_28339347 | 0.30 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
370 |
0.89 |
| chr20_39766624_39767421 | 0.29 |
RP1-1J6.2 |
|
379 |
0.79 |
| chr11_61485680_61486023 | 0.28 |
MYRF |
myelin regulatory factor |
34270 |
0.11 |
| chr1_20812928_20813453 | 0.28 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
477 |
0.82 |
| chr10_80166967_80167185 | 0.28 |
ENSG00000201393 |
. |
39812 |
0.22 |
| chr13_43148594_43148878 | 0.28 |
TNFSF11 |
tumor necrosis factor (ligand) superfamily, member 11 |
376 |
0.93 |
| chr1_23750455_23751189 | 0.28 |
TCEA3 |
transcription elongation factor A (SII), 3 |
378 |
0.83 |
| chr14_23306914_23307085 | 0.28 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
478 |
0.61 |
| chr14_23821578_23822379 | 0.28 |
SLC22A17 |
solute carrier family 22, member 17 |
91 |
0.91 |
| chr7_130572613_130572896 | 0.27 |
ENSG00000226380 |
. |
10456 |
0.27 |
| chr18_7117321_7117760 | 0.27 |
LAMA1 |
laminin, alpha 1 |
273 |
0.94 |
| chr10_60235657_60235933 | 0.27 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
37105 |
0.2 |
| chr12_59313444_59313663 | 0.27 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
226 |
0.87 |
| chr1_33219625_33220407 | 0.27 |
KIAA1522 |
KIAA1522 |
394 |
0.82 |
| chr2_177502027_177502943 | 0.27 |
ENSG00000252027 |
. |
26919 |
0.25 |
| chr9_97681675_97681826 | 0.26 |
RP11-54O15.3 |
|
15478 |
0.17 |
| chr2_129166329_129166512 | 0.26 |
ENSG00000238379 |
. |
36352 |
0.19 |
| chr10_17496627_17496975 | 0.26 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
472 |
0.84 |
| chr7_150822281_150822432 | 0.26 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
7060 |
0.11 |
| chr8_77594904_77595092 | 0.26 |
ZFHX4 |
zinc finger homeobox 4 |
236 |
0.67 |
| chr19_8428717_8428931 | 0.26 |
ANGPTL4 |
angiopoietin-like 4 |
211 |
0.87 |
| chr11_118492062_118492969 | 0.25 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
14157 |
0.11 |
| chr3_127175696_127176137 | 0.25 |
TPRA1 |
transmembrane protein, adipocyte asscociated 1 |
123364 |
0.05 |
| chr19_54982330_54982580 | 0.25 |
CDC42EP5 |
CDC42 effector protein (Rho GTPase binding) 5 |
1956 |
0.17 |
| chr13_36704716_36705075 | 0.25 |
DCLK1 |
doublecortin-like kinase 1 |
548 |
0.84 |
| chr15_29967513_29967692 | 0.25 |
RP11-680F8.1 |
|
408 |
0.87 |
| chr4_176568169_176568320 | 0.25 |
GPM6A |
glycoprotein M6A |
140294 |
0.05 |
| chr7_193532_193958 | 0.25 |
AC093627.12 |
|
435 |
0.67 |
| chr2_101436870_101437355 | 0.25 |
NPAS2 |
neuronal PAS domain protein 2 |
375 |
0.86 |
| chr17_70587270_70588900 | 0.24 |
ENSG00000200783 |
. |
72206 |
0.12 |
| chrX_149530380_149530720 | 0.24 |
MAMLD1 |
mastermind-like domain containing 1 |
1001 |
0.68 |
| chr10_123779225_123779376 | 0.24 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
2151 |
0.37 |
| chr10_29264874_29265025 | 0.24 |
ENSG00000199402 |
. |
101513 |
0.08 |
| chr3_192233089_192233240 | 0.24 |
FGF12-AS2 |
FGF12 antisense RNA 2 |
353 |
0.83 |
| chr22_44727371_44727573 | 0.24 |
KIAA1644 |
KIAA1644 |
18741 |
0.22 |
| chr21_38119758_38120093 | 0.23 |
SIM2 |
single-minded family bHLH transcription factor 2 |
35059 |
0.15 |
| chr10_135148565_135149526 | 0.23 |
CALY |
calcyon neuron-specific vesicular protein |
1331 |
0.22 |
| chr1_113286335_113286536 | 0.23 |
FAM19A3 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
23236 |
0.11 |
| chr1_210484700_210484949 | 0.23 |
HHAT |
hedgehog acyltransferase |
16772 |
0.23 |
| chr3_8671283_8671434 | 0.23 |
SSUH2 |
ssu-2 homolog (C. elegans) |
15126 |
0.15 |
| chr3_36422437_36422689 | 0.23 |
STAC |
SH3 and cysteine rich domain |
498 |
0.89 |
| chr5_95767258_95767555 | 0.23 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
458 |
0.88 |
| chrX_146273220_146273371 | 0.23 |
ENSG00000216171 |
. |
1990 |
0.19 |
| chr12_53612888_53613255 | 0.23 |
RARG |
retinoic acid receptor, gamma |
972 |
0.4 |
| chr19_38714929_38715117 | 0.23 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
133 |
0.92 |
| chr2_2650168_2650319 | 0.22 |
MYT1L |
myelin transcription factor 1-like |
315277 |
0.01 |
| chr19_17797735_17797934 | 0.22 |
UNC13A |
unc-13 homolog A (C. elegans) |
1174 |
0.41 |
| chr19_45888782_45889257 | 0.22 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
143 |
0.93 |
| chr9_100849786_100850488 | 0.22 |
TRIM14 |
tripartite motif containing 14 |
4706 |
0.2 |
| chr3_159756658_159757107 | 0.22 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
23071 |
0.18 |
| chr4_148693853_148694244 | 0.22 |
ENSG00000264274 |
. |
9698 |
0.18 |
| chr11_101918104_101918467 | 0.22 |
C11orf70 |
chromosome 11 open reading frame 70 |
96 |
0.96 |
| chr8_28347493_28348400 | 0.21 |
FBXO16 |
F-box protein 16 |
111 |
0.95 |
| chr4_2403656_2404488 | 0.21 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
4320 |
0.21 |
| chr10_830663_830851 | 0.21 |
LARP4B |
La ribonucleoprotein domain family, member 4B |
33029 |
0.18 |
| chr22_46933754_46934840 | 0.21 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
1230 |
0.46 |
| chr16_29231551_29231794 | 0.21 |
RP11-231C14.6 |
|
91999 |
0.06 |
| chr1_99470479_99470698 | 0.21 |
LPPR5 |
Lipid phosphate phosphatase-related protein type 5 |
0 |
0.95 |
| chr4_154407741_154408009 | 0.21 |
KIAA0922 |
KIAA0922 |
20374 |
0.23 |
| chr17_41984921_41985550 | 0.21 |
MPP2 |
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
108 |
0.94 |
| chr20_57426029_57426307 | 0.20 |
GNAS-AS1 |
GNAS antisense RNA 1 |
210 |
0.89 |
| chrY_16636977_16637245 | 0.20 |
NLGN4Y |
neuroligin 4, Y-linked |
657 |
0.84 |
| chr7_107612091_107612242 | 0.20 |
ENSG00000238297 |
. |
28261 |
0.14 |
| chr19_40698095_40698531 | 0.20 |
MAP3K10 |
mitogen-activated protein kinase kinase kinase 10 |
662 |
0.59 |
| chr19_50356246_50356397 | 0.20 |
PTOV1 |
prostate tumor overexpressed 1 |
1342 |
0.13 |
| chr6_104981791_104982069 | 0.20 |
ENSG00000252944 |
. |
232767 |
0.02 |
| chr1_31380173_31380807 | 0.20 |
SDC3 |
syndecan 3 |
1118 |
0.44 |
| chr7_562042_562272 | 0.20 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
2224 |
0.31 |
| chr20_2110844_2110995 | 0.19 |
ENSG00000263452 |
. |
27008 |
0.18 |
| chr5_78985270_78986298 | 0.19 |
CMYA5 |
cardiomyopathy associated 5 |
84 |
0.98 |
| chr17_79093601_79093928 | 0.19 |
ENSG00000207736 |
. |
5409 |
0.1 |
| chr4_150999769_151000790 | 0.19 |
DCLK2 |
doublecortin-like kinase 2 |
99 |
0.98 |
| chr16_54321004_54321411 | 0.19 |
IRX3 |
iroquois homeobox 3 |
532 |
0.83 |
| chr8_1771644_1772131 | 0.19 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
255 |
0.93 |
| chr7_560180_560411 | 0.19 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
362 |
0.88 |
| chr7_73704189_73704652 | 0.19 |
CLIP2 |
CAP-GLY domain containing linker protein 2 |
615 |
0.73 |
| chr16_67433264_67433415 | 0.19 |
ZDHHC1 |
zinc finger, DHHC-type containing 1 |
1555 |
0.23 |
| chr14_37050333_37050591 | 0.19 |
NKX2-8 |
NK2 homeobox 8 |
1350 |
0.4 |
| chr2_130901985_130902795 | 0.19 |
CCDC74B |
coiled-coil domain containing 74B |
177 |
0.9 |
| chr8_494526_494820 | 0.19 |
TDRP |
testis development related protein |
164 |
0.97 |
| chr1_10926741_10927032 | 0.18 |
CASZ1 |
castor zinc finger 1 |
70179 |
0.09 |
| chr17_19374169_19374649 | 0.18 |
ENSG00000265335 |
. |
50817 |
0.08 |
| chr7_150812372_150812612 | 0.18 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
727 |
0.51 |
| chr22_16192781_16192941 | 0.18 |
LL22NC03-N64E9.1 |
|
11857 |
0.18 |
| chr1_210465594_210466382 | 0.18 |
HHAT |
hedgehog acyltransferase |
35608 |
0.17 |
| chr4_55096373_55096991 | 0.18 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
193 |
0.97 |
| chr2_73496654_73496805 | 0.18 |
FBXO41 |
F-box protein 41 |
1314 |
0.36 |
| chr16_85548809_85549111 | 0.18 |
ENSG00000264203 |
. |
73862 |
0.1 |
| chr2_191629077_191629228 | 0.18 |
AC006460.2 |
|
55710 |
0.13 |
| chr6_28522205_28522356 | 0.18 |
GPX5 |
glutathione peroxidase 5 (epididymal androgen-related protein) |
28491 |
0.14 |
| chr11_118791642_118792157 | 0.18 |
BCL9L |
B-cell CLL/lymphoma 9-like |
2286 |
0.14 |
| chr10_30346435_30346778 | 0.18 |
KIAA1462 |
KIAA1462 |
1847 |
0.5 |
| chr17_36621277_36621585 | 0.18 |
ARHGAP23 |
Rho GTPase activating protein 23 |
6749 |
0.17 |
| chr14_99655246_99655958 | 0.18 |
AL162151.4 |
|
30849 |
0.2 |
| chr15_33602886_33603116 | 0.18 |
RP11-489D6.2 |
|
141 |
0.53 |
| chr8_102168134_102168418 | 0.18 |
ENSG00000202360 |
. |
18089 |
0.22 |
| chr2_133427812_133428020 | 0.18 |
LYPD1 |
LY6/PLAUR domain containing 1 |
138 |
0.97 |
| chr15_57884211_57884584 | 0.18 |
GCOM1 |
GRINL1A complex locus 1 |
166 |
0.33 |
| chr16_2128784_2128935 | 0.18 |
TSC2 |
tuberous sclerosis 2 |
4332 |
0.07 |
| chr6_3248086_3248639 | 0.18 |
PSMG4 |
proteasome (prosome, macropain) assembly chaperone 4 |
10766 |
0.18 |
| chr19_39466589_39467071 | 0.18 |
FBXO17 |
F-box protein 17 |
280 |
0.84 |
| chr13_30168298_30169638 | 0.17 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
857 |
0.74 |
| chr2_27717607_27718081 | 0.17 |
FNDC4 |
fibronectin type III domain containing 4 |
268 |
0.82 |
| chr12_22093968_22094205 | 0.17 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
81 |
0.98 |
| chr6_80656707_80657785 | 0.17 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
51 |
0.98 |
| chr19_19006269_19006725 | 0.17 |
GDF1 |
growth differentiation factor 1 |
408 |
0.46 |
| chr3_125985987_125986785 | 0.17 |
ALDH1L1-AS2 |
ALDH1L1 antisense RNA 2 |
60288 |
0.1 |
| chr7_767439_768007 | 0.17 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
436 |
0.68 |
| chr4_14864468_14865022 | 0.17 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
139553 |
0.05 |
| chr5_170024412_170024773 | 0.17 |
CTC-265N9.1 |
|
83849 |
0.08 |
| chr8_79428088_79428454 | 0.17 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
103 |
0.98 |
| chr2_132439969_132440520 | 0.17 |
C2orf27A |
chromosome 2 open reading frame 27A |
39704 |
0.17 |
| chr11_8041404_8041600 | 0.17 |
TUB |
tubby bipartite transcription factor |
711 |
0.56 |
| chr16_66914963_66915191 | 0.17 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
567 |
0.61 |
| chr19_1748094_1748245 | 0.17 |
ONECUT3 |
one cut homeobox 3 |
4203 |
0.16 |
| chr16_88449218_88449497 | 0.17 |
ZNF469 |
zinc finger protein 469 |
44522 |
0.14 |
| chr11_69457709_69458273 | 0.16 |
CCND1 |
cyclin D1 |
2017 |
0.36 |
| chr11_62691064_62691268 | 0.16 |
CHRM1 |
cholinergic receptor, muscarinic 1 |
1887 |
0.2 |
| chr20_42955224_42955721 | 0.16 |
R3HDML |
R3H domain containing-like |
10154 |
0.11 |
| chrX_117957425_117957978 | 0.16 |
ZCCHC12 |
zinc finger, CCHC domain containing 12 |
52 |
0.98 |
| chr20_44034499_44035314 | 0.16 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
45 |
0.96 |
| chr19_49842468_49842681 | 0.16 |
CD37 |
CD37 molecule |
2102 |
0.15 |
| chr4_52904501_52904984 | 0.16 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
94 |
0.97 |
| chr9_34651822_34652525 | 0.16 |
IL11RA |
interleukin 11 receptor, alpha |
16 |
0.94 |
| chr13_102068214_102068409 | 0.16 |
NALCN |
sodium leak channel, non-selective |
395 |
0.91 |
| chrX_12156590_12157176 | 0.16 |
FRMPD4 |
FERM and PDZ domain containing 4 |
298 |
0.9 |
| chr4_7940010_7940502 | 0.16 |
AC097381.1 |
|
472 |
0.75 |
| chr5_137801989_137802208 | 0.16 |
EGR1 |
early growth response 1 |
919 |
0.54 |
| chr1_225117342_225117684 | 0.16 |
DNAH14 |
dynein, axonemal, heavy chain 14 |
129 |
0.98 |
| chr9_17579467_17579684 | 0.16 |
SH3GL2 |
SH3-domain GRB2-like 2 |
454 |
0.91 |
| chr19_50836422_50837123 | 0.16 |
KCNC3 |
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
0 |
0.95 |
| chr1_82268816_82269203 | 0.16 |
LPHN2 |
latrophilin 2 |
2927 |
0.41 |
| chr11_35641402_35641625 | 0.16 |
FJX1 |
four jointed box 1 (Drosophila) |
1778 |
0.42 |
| chr4_187476503_187476829 | 0.16 |
MTNR1A |
melatonin receptor 1A |
55 |
0.7 |
| chr20_10197984_10198249 | 0.16 |
SNAP25 |
synaptosomal-associated protein, 25kDa |
1362 |
0.42 |
| chr1_153581254_153581448 | 0.16 |
S100A16 |
S100 calcium binding protein A16 |
462 |
0.62 |
| chr2_56151103_56151254 | 0.16 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
96 |
0.97 |
| chr6_85474025_85474422 | 0.16 |
TBX18 |
T-box 18 |
14 |
0.99 |
| chr16_54962446_54962612 | 0.16 |
IRX5 |
iroquois homeobox 5 |
2245 |
0.47 |
| chr6_28227027_28227486 | 0.16 |
NKAPL |
NFKB activating protein-like |
158 |
0.61 |
| chr6_33589286_33590289 | 0.15 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
626 |
0.65 |
| chr19_39360695_39361085 | 0.15 |
CTC-360G5.6 |
|
91 |
0.64 |
| chr11_65158052_65158759 | 0.15 |
FRMD8 |
FERM domain containing 8 |
4006 |
0.13 |
| chr13_22257522_22257673 | 0.15 |
FGF9 |
fibroblast growth factor 9 |
12075 |
0.25 |
| chr19_8204803_8204954 | 0.15 |
FBN3 |
fibrillin 3 |
7772 |
0.19 |
| chr12_13045017_13045168 | 0.15 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
184 |
0.93 |
| chr7_558289_558902 | 0.15 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
436 |
0.84 |
| chr19_39904322_39904875 | 0.15 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
146 |
0.89 |
| chr17_70026380_70026708 | 0.15 |
AC007461.1 |
Uncharacterized protein |
9620 |
0.31 |
| chr6_133562881_133563193 | 0.15 |
EYA4 |
eyes absent homolog 4 (Drosophila) |
268 |
0.95 |
| chr4_5889500_5889651 | 0.15 |
CRMP1 |
collapsin response mediator protein 1 |
633 |
0.79 |
| chr18_12287984_12288154 | 0.15 |
ENSG00000199702 |
. |
16124 |
0.16 |
| chrX_52927117_52927336 | 0.15 |
FAM156B |
family with sequence similarity 156, member B |
350 |
0.88 |
| chr9_98279334_98279613 | 0.15 |
PTCH1 |
patched 1 |
134 |
0.95 |
| chr19_40972554_40972858 | 0.15 |
SPTBN4 |
spectrin, beta, non-erythrocytic 4 |
420 |
0.65 |
| chr8_22085028_22085274 | 0.15 |
PHYHIP |
phytanoyl-CoA 2-hydroxylase interacting protein |
55 |
0.96 |
| chr19_42456013_42456185 | 0.15 |
RABAC1 |
Rab acceptor 1 (prenylated) |
6967 |
0.14 |
| chrX_10141481_10141728 | 0.15 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
15032 |
0.23 |
| chr11_62312562_62314176 | 0.15 |
RP11-864I4.4 |
|
102 |
0.64 |
| chr3_184056155_184056581 | 0.15 |
FAM131A |
family with sequence similarity 131, member A |
249 |
0.84 |
| chr7_1600163_1600314 | 0.15 |
TMEM184A |
transmembrane protein 184A |
219 |
0.92 |
| chr13_23733598_23733940 | 0.15 |
ENSG00000207157 |
. |
6944 |
0.23 |
| chr16_3241653_3241969 | 0.15 |
AJ003147.9 |
|
3599 |
0.12 |
| chr5_131593477_131594185 | 0.15 |
PDLIM4 |
PDZ and LIM domain 4 |
431 |
0.81 |
| chr1_38259533_38260044 | 0.15 |
MANEAL |
mannosidase, endo-alpha-like |
233 |
0.86 |
| chr22_30641646_30641949 | 0.15 |
RP1-102K2.8 |
|
187 |
0.85 |
| chr6_36567708_36568239 | 0.15 |
SRSF3 |
serine/arginine-rich splicing factor 3 |
5828 |
0.16 |
| chr19_56576699_56576850 | 0.15 |
ZNF787 |
zinc finger protein 787 |
38079 |
0.1 |
| chr5_137609759_137610630 | 0.15 |
GFRA3 |
GDNF family receptor alpha 3 |
60 |
0.96 |
| chr9_117880223_117880437 | 0.15 |
TNC |
tenascin C |
148 |
0.97 |
| chr17_4902045_4902282 | 0.15 |
KIF1C |
kinesin family member 1C |
914 |
0.26 |
| chr5_124827830_124828314 | 0.15 |
ENSG00000222107 |
. |
141248 |
0.05 |
| chr17_62971851_62972399 | 0.15 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
2490 |
0.23 |
| chr5_139283809_139284341 | 0.15 |
NRG2 |
neuregulin 2 |
93 |
0.98 |
| chr7_138720144_138720977 | 0.15 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
215 |
0.95 |
| chr5_134824456_134824665 | 0.15 |
TIFAB |
TRAF-interacting protein with forkhead-associated domain, family member B |
36471 |
0.11 |
| chr1_20811158_20811808 | 0.14 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
1230 |
0.48 |
| chr16_65157518_65157704 | 0.14 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
1343 |
0.63 |
| chr11_70244216_70245100 | 0.14 |
CTTN |
cortactin |
11 |
0.53 |
| chr17_9142709_9143234 | 0.14 |
RP11-85B7.4 |
|
43520 |
0.15 |
| chr22_18958500_18958651 | 0.14 |
DGCR5 |
DiGeorge syndrome critical region gene 5 (non-protein coding) |
479 |
0.78 |
| chr5_95767628_95767788 | 0.14 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
156 |
0.97 |
| chr18_74271196_74271347 | 0.14 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
30397 |
0.18 |
| chr1_19282077_19282398 | 0.14 |
IFFO2 |
intermediate filament family orphan 2 |
45 |
0.98 |
| chr10_74544262_74544615 | 0.14 |
RP11-354E23.5 |
|
18120 |
0.19 |
| chr1_40098862_40099023 | 0.14 |
RP1-144F13.3 |
|
146 |
0.66 |
| chr7_158823123_158823874 | 0.14 |
ENSG00000231419 |
. |
22386 |
0.25 |
| chr8_17354234_17354457 | 0.14 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
252 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 0.5 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 0.4 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.1 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.2 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) |
| 0.1 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.3 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.1 | GO:0072110 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0072239 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.0 | 0.3 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.3 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0090185 | negative regulation of kidney development(GO:0090185) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0033240 | positive regulation of cellular amine metabolic process(GO:0033240) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.2 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.3 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.0 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.0 | GO:1901722 | cell proliferation involved in metanephros development(GO:0072203) regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.3 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.3 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0060033 | Mullerian duct regression(GO:0001880) anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.0 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) positive regulation of cardiocyte differentiation(GO:1905209) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0019614 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.0 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0071599 | otic vesicle formation(GO:0030916) otic vesicle development(GO:0071599) otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:0048799 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.0 | 0.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:0043092 | L-amino acid import(GO:0043092) |
| 0.0 | 0.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 0.0 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0042596 | behavioral fear response(GO:0001662) fear response(GO:0042596) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.0 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.0 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.4 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0099528 | dopamine neurotransmitter receptor activity(GO:0004952) G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |