Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
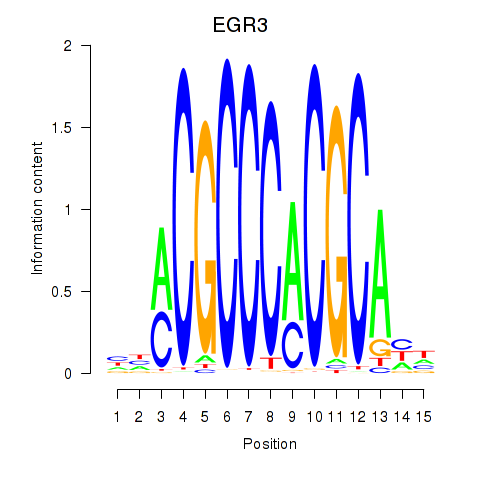
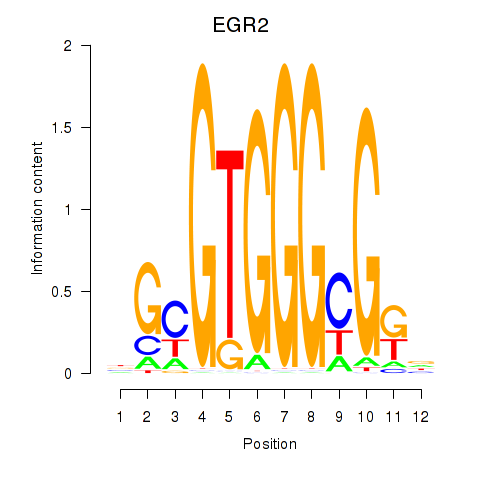
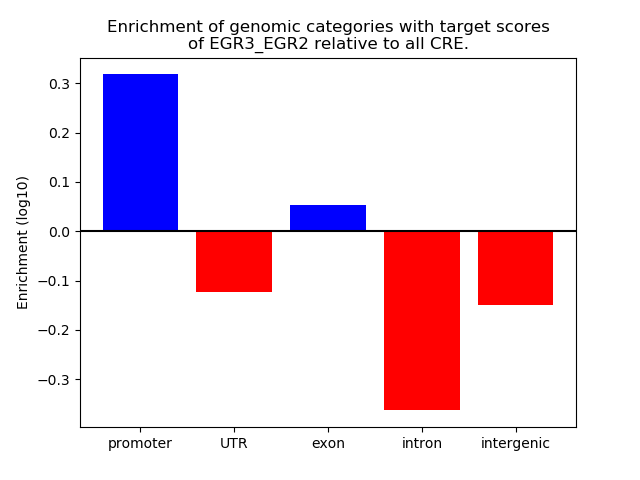
Results for EGR3_EGR2
Z-value: 0.55
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR3
|
ENSG00000179388.8 | early growth response 3 |
|
EGR2
|
ENSG00000122877.9 | early growth response 2 |
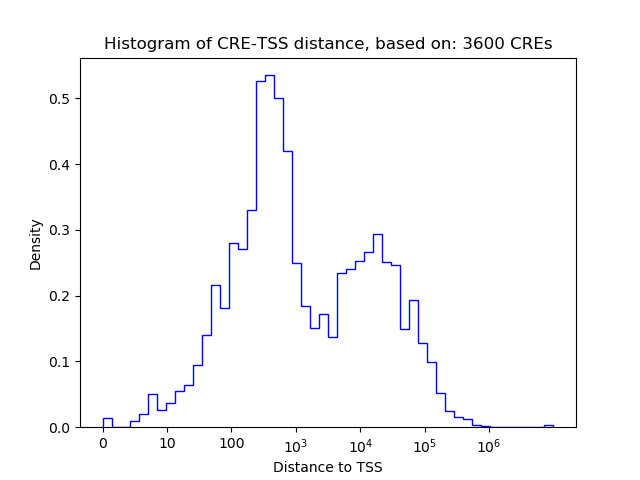
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_64576967_64577230 | EGR2 | 972 | 0.568878 | -0.66 | 5.5e-02 | Click! |
| chr10_64577478_64577629 | EGR2 | 1374 | 0.440127 | -0.61 | 7.9e-02 | Click! |
| chr10_64645936_64646087 | EGR2 | 67084 | 0.111874 | 0.49 | 1.8e-01 | Click! |
| chr10_64577295_64577446 | EGR2 | 1244 | 0.474933 | -0.49 | 1.8e-01 | Click! |
| chr10_64576654_64576951 | EGR2 | 676 | 0.707493 | -0.46 | 2.2e-01 | Click! |
| chr8_22552129_22552945 | EGR3 | 1722 | 0.257808 | -0.67 | 4.8e-02 | Click! |
| chr8_22551556_22552015 | EGR3 | 970 | 0.435958 | -0.60 | 8.7e-02 | Click! |
| chr8_22572743_22572905 | EGR3 | 22009 | 0.124907 | 0.58 | 9.8e-02 | Click! |
| chr8_22551057_22551465 | EGR3 | 446 | 0.740990 | -0.58 | 1.0e-01 | Click! |
| chr8_22576706_22576857 | EGR3 | 25966 | 0.121391 | -0.54 | 1.4e-01 | Click! |
Activity of the EGR3_EGR2 motif across conditions
Conditions sorted by the z-value of the EGR3_EGR2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
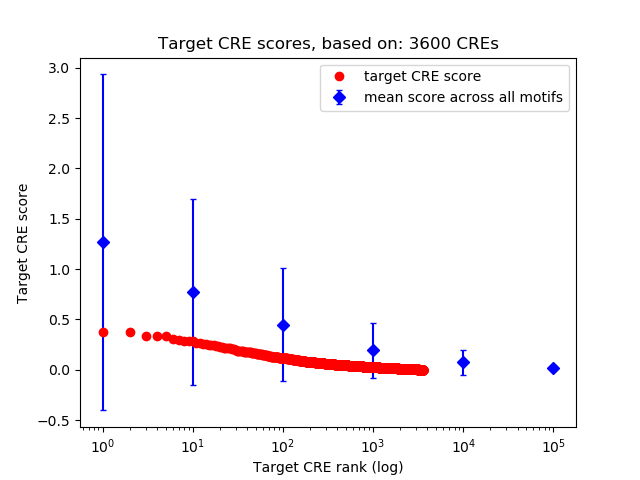
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_49347136_49348386 | 0.37 |
PARD6B |
par-6 family cell polarity regulator beta |
320 |
0.89 |
| chr7_12755011_12755366 | 0.37 |
ENSG00000199470 |
. |
14674 |
0.23 |
| chr21_45139141_45140358 | 0.34 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
756 |
0.66 |
| chr9_100069234_100069583 | 0.34 |
CCDC180 |
coiled-coil domain containing 180 |
533 |
0.83 |
| chr4_150999769_151000790 | 0.33 |
DCLK2 |
doublecortin-like kinase 2 |
99 |
0.98 |
| chr5_63461313_63462499 | 0.30 |
RNF180 |
ring finger protein 180 |
83 |
0.99 |
| chr2_242497891_242499073 | 0.30 |
BOK-AS1 |
BOK antisense RNA 1 |
90 |
0.79 |
| chr1_9189517_9189890 | 0.29 |
GPR157 |
G protein-coupled receptor 157 |
474 |
0.79 |
| chr1_33336564_33337052 | 0.29 |
FNDC5 |
fibronectin type III domain containing 5 |
394 |
0.8 |
| chr19_15311959_15312270 | 0.28 |
NOTCH3 |
notch 3 |
322 |
0.87 |
| chr4_121842864_121843514 | 0.27 |
PRDM5 |
PR domain containing 5 |
815 |
0.75 |
| chr19_56988243_56988602 | 0.26 |
ZNF667 |
zinc finger protein 667 |
65 |
0.96 |
| chr1_86042054_86042529 | 0.26 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
1642 |
0.36 |
| chr1_159750747_159751482 | 0.26 |
DUSP23 |
dual specificity phosphatase 23 |
321 |
0.85 |
| chr1_244012088_244013269 | 0.25 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
752 |
0.75 |
| chr17_57408824_57410302 | 0.25 |
YPEL2 |
yippee-like 2 (Drosophila) |
500 |
0.78 |
| chr14_91790220_91790791 | 0.24 |
ENSG00000265856 |
. |
9552 |
0.21 |
| chr5_153990189_153990659 | 0.24 |
ENSG00000264760 |
. |
14792 |
0.21 |
| chr17_8602020_8602357 | 0.24 |
CCDC42 |
coiled-coil domain containing 42 |
45917 |
0.12 |
| chr2_946282_946607 | 0.23 |
SNTG2 |
syntrophin, gamma 2 |
110 |
0.86 |
| chr16_402538_402771 | 0.23 |
AXIN1 |
axin 1 |
5 |
0.96 |
| chr15_80696207_80696642 | 0.22 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
269 |
0.71 |
| chr3_196729783_196730213 | 0.22 |
MFI2-AS1 |
MFI2 antisense RNA 1 |
5 |
0.97 |
| chr11_64512499_64513555 | 0.21 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
99 |
0.95 |
| chr10_49812568_49813319 | 0.21 |
ARHGAP22 |
Rho GTPase activating protein 22 |
54 |
0.98 |
| chr4_3709787_3709938 | 0.21 |
ADRA2C |
adrenoceptor alpha 2C |
58213 |
0.14 |
| chr6_28175094_28175274 | 0.21 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
17480 |
0.12 |
| chr1_31380173_31380807 | 0.21 |
SDC3 |
syndecan 3 |
1118 |
0.44 |
| chr2_177502027_177502943 | 0.20 |
ENSG00000252027 |
. |
26919 |
0.25 |
| chr12_63211718_63211943 | 0.19 |
ENSG00000200296 |
. |
32851 |
0.21 |
| chr7_127672748_127672947 | 0.19 |
LRRC4 |
leucine rich repeat containing 4 |
687 |
0.76 |
| chr4_79472640_79473562 | 0.19 |
ANXA3 |
annexin A3 |
20 |
0.99 |
| chr8_145925356_145925968 | 0.19 |
AF186192.5 |
|
5257 |
0.15 |
| chr1_3816010_3816785 | 0.19 |
C1orf174 |
chromosome 1 open reading frame 174 |
452 |
0.8 |
| chr12_63025567_63026419 | 0.18 |
ENSG00000238475 |
. |
18962 |
0.14 |
| chr2_26624476_26625174 | 0.18 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
41 |
0.98 |
| chr1_1370771_1371229 | 0.18 |
VWA1 |
von Willebrand factor A domain containing 1 |
51 |
0.93 |
| chr12_6937518_6938631 | 0.18 |
LEPREL2 |
leprecan-like 2 |
502 |
0.55 |
| chr1_205512616_205512824 | 0.18 |
ENSG00000253097 |
. |
23124 |
0.13 |
| chr16_88857124_88857275 | 0.18 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
5580 |
0.1 |
| chr4_14864468_14865022 | 0.18 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
139553 |
0.05 |
| chr6_146919852_146920086 | 0.17 |
ADGB |
androglobin |
132 |
0.97 |
| chr3_37903731_37903951 | 0.17 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
176 |
0.83 |
| chr16_402920_403077 | 0.17 |
AXIN1 |
axin 1 |
339 |
0.79 |
| chr6_166076326_166076646 | 0.17 |
PDE10A |
phosphodiesterase 10A |
898 |
0.71 |
| chr7_100081068_100081412 | 0.17 |
ENSG00000266372 |
. |
226 |
0.55 |
| chr7_28450564_28450725 | 0.17 |
CREB5 |
cAMP responsive element binding protein 5 |
1500 |
0.57 |
| chr4_134072879_134073697 | 0.17 |
PCDH10 |
protocadherin 10 |
2818 |
0.44 |
| chr9_98279334_98279613 | 0.17 |
PTCH1 |
patched 1 |
134 |
0.95 |
| chr5_158636669_158637140 | 0.16 |
CTB-11I22.1 |
|
25 |
0.66 |
| chr2_8818889_8820001 | 0.16 |
AC011747.7 |
|
259 |
0.7 |
| chr3_141379125_141379497 | 0.16 |
RNF7 |
ring finger protein 7 |
77735 |
0.09 |
| chr2_167350846_167351007 | 0.16 |
SCN7A |
sodium channel, voltage-gated, type VII, alpha subunit |
169 |
0.97 |
| chr7_193532_193958 | 0.16 |
AC093627.12 |
|
435 |
0.67 |
| chr18_2848081_2848464 | 0.16 |
EMILIN2 |
elastin microfibril interfacer 2 |
1244 |
0.42 |
| chr19_38714929_38715117 | 0.15 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
133 |
0.92 |
| chr1_227058397_227059047 | 0.15 |
PSEN2 |
presenilin 2 (Alzheimer disease 4) |
243 |
0.94 |
| chr17_7905804_7906020 | 0.15 |
GUCY2D |
guanylate cyclase 2D, membrane (retina-specific) |
0 |
0.96 |
| chr18_12287984_12288154 | 0.15 |
ENSG00000199702 |
. |
16124 |
0.16 |
| chr19_38878742_38879193 | 0.15 |
SPRED3 |
sprouty-related, EVH1 domain containing 3 |
94 |
0.68 |
| chr9_100849786_100850488 | 0.15 |
TRIM14 |
tripartite motif containing 14 |
4706 |
0.2 |
| chr3_46924906_46925224 | 0.15 |
PTH1R |
parathyroid hormone 1 receptor |
218 |
0.88 |
| chr1_19600410_19601051 | 0.15 |
AKR7L |
aldo-keto reductase family 7-like |
162 |
0.92 |
| chr17_60885608_60885921 | 0.14 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
59 |
0.98 |
| chr10_13500683_13500834 | 0.14 |
BEND7 |
BEN domain containing 7 |
22315 |
0.16 |
| chr1_200003972_200004421 | 0.14 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
7185 |
0.18 |
| chr18_32073627_32073820 | 0.14 |
DTNA |
dystrobrevin, alpha |
144 |
0.98 |
| chr2_27717607_27718081 | 0.14 |
FNDC4 |
fibronectin type III domain containing 4 |
268 |
0.82 |
| chr19_15442836_15443133 | 0.13 |
BRD4 |
bromodomain containing 4 |
266 |
0.91 |
| chr8_1921192_1922084 | 0.13 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
406 |
0.9 |
| chr4_187476503_187476829 | 0.13 |
MTNR1A |
melatonin receptor 1A |
55 |
0.7 |
| chr4_55096373_55096991 | 0.13 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
193 |
0.97 |
| chr19_23258281_23258533 | 0.13 |
ZNF730 |
zinc finger protein 730 |
395 |
0.9 |
| chr22_20003723_20004755 | 0.13 |
ARVCF |
armadillo repeat gene deleted in velocardiofacial syndrome |
92 |
0.75 |
| chr10_103589679_103590635 | 0.13 |
KCNIP2 |
Kv channel interacting protein 2 |
38 |
0.97 |
| chr9_115875359_115875603 | 0.13 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
37741 |
0.15 |
| chr2_103235205_103235939 | 0.13 |
SLC9A2 |
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
594 |
0.82 |
| chr2_105489505_105490071 | 0.13 |
LINC01159 |
long intergenic non-protein coding RNA 1159 |
735 |
0.58 |
| chrX_21958323_21959013 | 0.13 |
SMS |
spermine synthase |
23 |
0.98 |
| chr10_116503730_116503881 | 0.13 |
ABLIM1 |
actin binding LIM protein 1 |
59391 |
0.13 |
| chr17_15653076_15653227 | 0.13 |
AC005324.6 |
|
645 |
0.65 |
| chr6_166721057_166722024 | 0.13 |
PRR18 |
proline rich 18 |
331 |
0.9 |
| chr11_118478413_118479735 | 0.13 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
716 |
0.54 |
| chrX_37544746_37545007 | 0.13 |
XK |
X-linked Kx blood group (McLeod syndrome) |
136 |
0.98 |
| chr19_36207272_36208543 | 0.13 |
KMT2B |
Histone-lysine N-methyltransferase 2B |
1014 |
0.26 |
| chr21_40032373_40032758 | 0.13 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
26 |
0.99 |
| chr4_108745530_108746004 | 0.12 |
SGMS2 |
sphingomyelin synthase 2 |
48 |
0.98 |
| chr9_115873523_115873775 | 0.12 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
39573 |
0.15 |
| chr16_88219567_88219948 | 0.12 |
BANP |
BTG3 associated nuclear protein |
216133 |
0.02 |
| chr11_57282891_57283496 | 0.12 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
34 |
0.95 |
| chr16_88449218_88449497 | 0.12 |
ZNF469 |
zinc finger protein 469 |
44522 |
0.14 |
| chr20_32899126_32899277 | 0.12 |
AHCY |
adenosylhomocysteinase |
407 |
0.81 |
| chr14_85999757_86000024 | 0.12 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
3318 |
0.3 |
| chrX_39949292_39950247 | 0.12 |
BCOR |
BCL6 corepressor |
6887 |
0.33 |
| chrX_39964334_39965624 | 0.12 |
BCOR |
BCL6 corepressor |
8323 |
0.32 |
| chr9_130639317_130639975 | 0.12 |
AK1 |
adenylate kinase 1 |
376 |
0.71 |
| chr8_61821706_61823076 | 0.12 |
RP11-33I11.2 |
|
100226 |
0.08 |
| chr6_762027_762197 | 0.12 |
EXOC2 |
exocyst complex component 2 |
68995 |
0.13 |
| chr12_29936821_29937292 | 0.12 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
226 |
0.96 |
| chr4_186130212_186131033 | 0.12 |
KIAA1430 |
KIAA1430 |
36 |
0.92 |
| chr17_18061433_18061699 | 0.11 |
MYO15A |
myosin XVA |
56 |
0.96 |
| chr11_122311916_122312418 | 0.11 |
ENSG00000252776 |
. |
7751 |
0.23 |
| chr3_187769593_187769744 | 0.11 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
101404 |
0.07 |
| chr2_99757819_99758492 | 0.11 |
C2ORF15 |
Uncharacterized protein C2orf15 |
58 |
0.58 |
| chr12_75728365_75728595 | 0.11 |
GLIPR1L1 |
GLI pathogenesis-related 1 like 1 |
17 |
0.98 |
| chr7_29846053_29846716 | 0.11 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
282 |
0.94 |
| chr11_61485680_61486023 | 0.11 |
MYRF |
myelin regulatory factor |
34270 |
0.11 |
| chr19_10982465_10983821 | 0.11 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
764 |
0.53 |
| chr20_30196082_30196710 | 0.11 |
ENSG00000264395 |
. |
1407 |
0.28 |
| chr7_19152184_19152434 | 0.11 |
AC003986.6 |
|
212 |
0.92 |
| chr12_54762715_54762975 | 0.11 |
GPR84 |
G protein-coupled receptor 84 |
4574 |
0.1 |
| chr9_132382776_132383013 | 0.11 |
C9orf50 |
chromosome 9 open reading frame 50 |
161 |
0.92 |
| chr9_17579467_17579684 | 0.11 |
SH3GL2 |
SH3-domain GRB2-like 2 |
454 |
0.91 |
| chr14_23706832_23707104 | 0.11 |
C14orf164 |
chromosome 14 open reading frame 164 |
2587 |
0.16 |
| chr22_50919615_50920637 | 0.11 |
ADM2 |
adrenomedullin 2 |
27 |
0.94 |
| chrX_138774003_138774873 | 0.11 |
MCF2 |
MCF.2 cell line derived transforming sequence |
123 |
0.97 |
| chr6_133089264_133089460 | 0.11 |
VNN2 |
vanin 2 |
4764 |
0.14 |
| chr11_13690295_13691391 | 0.11 |
FAR1-IT1 |
FAR1 intronic transcript 1 (non-protein coding) |
31 |
0.92 |
| chr16_29231551_29231794 | 0.11 |
RP11-231C14.6 |
|
91999 |
0.06 |
| chr15_40697977_40698740 | 0.11 |
IVD |
isovaleryl-CoA dehydrogenase |
83 |
0.94 |
| chr1_225839301_225840394 | 0.11 |
ENAH |
enabled homolog (Drosophila) |
571 |
0.78 |
| chr19_49866897_49867069 | 0.11 |
DKKL1 |
dickkopf-like 1 |
4 |
0.93 |
| chr2_129166329_129166512 | 0.10 |
ENSG00000238379 |
. |
36352 |
0.19 |
| chrX_48814395_48814616 | 0.10 |
OTUD5 |
OTU domain containing 5 |
54 |
0.94 |
| chr7_145813028_145813349 | 0.10 |
CNTNAP2 |
contactin associated protein-like 2 |
265 |
0.94 |
| chr1_95286002_95286302 | 0.10 |
SLC44A3 |
solute carrier family 44, member 3 |
25 |
0.98 |
| chr3_39093427_39094272 | 0.10 |
WDR48 |
WD repeat domain 48 |
57 |
0.97 |
| chr16_54318004_54318459 | 0.10 |
IRX3 |
iroquois homeobox 3 |
1536 |
0.47 |
| chr8_637614_638228 | 0.10 |
ERICH1 |
glutamate-rich 1 |
14264 |
0.22 |
| chr6_1619961_1620659 | 0.10 |
FOXC1 |
forkhead box C1 |
9629 |
0.3 |
| chr10_81194638_81194815 | 0.10 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
9249 |
0.21 |
| chr18_56029142_56029527 | 0.10 |
RP11-845C23.2 |
|
9464 |
0.19 |
| chr1_27320396_27320765 | 0.10 |
TRNP1 |
TMF1-regulated nuclear protein 1 |
230 |
0.91 |
| chr8_125673018_125673304 | 0.10 |
MTSS1 |
metastasis suppressor 1 |
66696 |
0.11 |
| chr7_94025506_94025990 | 0.10 |
COL1A2 |
collagen, type I, alpha 2 |
1875 |
0.46 |
| chr7_93520142_93520396 | 0.10 |
TFPI2 |
tissue factor pathway inhibitor 2 |
34 |
0.5 |
| chr12_111843881_111845308 | 0.10 |
SH2B3 |
SH2B adaptor protein 3 |
842 |
0.62 |
| chrX_40035041_40035983 | 0.10 |
BCOR |
BCL6 corepressor |
1061 |
0.68 |
| chrX_117861722_117862389 | 0.10 |
IL13RA1 |
interleukin 13 receptor, alpha 1 |
490 |
0.84 |
| chrX_101186111_101187162 | 0.10 |
ZMAT1 |
zinc finger, matrin-type 1 |
101 |
0.97 |
| chr21_37529152_37530122 | 0.10 |
DOPEY2 |
dopey family member 2 |
557 |
0.57 |
| chr15_100272658_100273585 | 0.10 |
LYSMD4 |
LysM, putative peptidoglycan-binding, domain containing 4 |
423 |
0.79 |
| chr6_166421798_166421999 | 0.10 |
ENSG00000252196 |
. |
66069 |
0.12 |
| chr6_164203543_164203694 | 0.09 |
ENSG00000266128 |
. |
57975 |
0.17 |
| chr21_47401799_47402727 | 0.09 |
COL6A1 |
collagen, type VI, alpha 1 |
612 |
0.75 |
| chr19_48996311_48996470 | 0.09 |
LMTK3 |
lemur tyrosine kinase 3 |
18671 |
0.09 |
| chr1_47184939_47185223 | 0.09 |
EFCAB14 |
EF-hand calcium binding domain 14 |
257 |
0.91 |
| chr8_71005594_71005799 | 0.09 |
ENSG00000212473 |
. |
15429 |
0.2 |
| chr2_96012468_96013004 | 0.09 |
KCNIP3 |
Kv channel interacting protein 3, calsenilin |
260 |
0.94 |
| chr2_204193085_204194072 | 0.09 |
ABI2 |
abl-interactor 2 |
371 |
0.48 |
| chr17_13505362_13505866 | 0.09 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
370 |
0.91 |
| chr10_133793290_133793613 | 0.09 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
1976 |
0.41 |
| chrX_134556811_134556962 | 0.09 |
ENSG00000201440 |
. |
5324 |
0.25 |
| chr3_169376087_169376335 | 0.09 |
MECOM |
MDS1 and EVI1 complex locus |
4967 |
0.2 |
| chr22_39377484_39377635 | 0.09 |
APOBEC3B |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
793 |
0.51 |
| chr5_102594979_102595784 | 0.09 |
C5orf30 |
chromosome 5 open reading frame 30 |
256 |
0.94 |
| chr15_33010201_33010591 | 0.09 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
191 |
0.95 |
| chr9_140335891_140336276 | 0.09 |
ENTPD8 |
ectonucleoside triphosphate diphosphohydrolase 8 |
182 |
0.91 |
| chr19_56159091_56160130 | 0.09 |
CCDC106 |
coiled-coil domain containing 106 |
11 |
0.93 |
| chr22_29469254_29470042 | 0.09 |
KREMEN1 |
kringle containing transmembrane protein 1 |
529 |
0.72 |
| chr7_97912986_97913801 | 0.09 |
BRI3 |
brain protein I3 |
2406 |
0.26 |
| chr10_81206278_81206598 | 0.09 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
1055 |
0.56 |
| chr1_205648469_205649627 | 0.09 |
SLC45A3 |
solute carrier family 45, member 3 |
539 |
0.74 |
| chr3_170075598_170076731 | 0.09 |
SKIL |
SKI-like oncogene |
642 |
0.78 |
| chr10_38893897_38894048 | 0.09 |
LINC00999 |
long intergenic non-protein coding RNA 999 |
176898 |
0.03 |
| chr7_107641976_107642307 | 0.09 |
LAMB1 |
laminin, beta 1 |
331 |
0.84 |
| chr9_15510511_15510905 | 0.09 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
281 |
0.9 |
| chr7_128828157_128829026 | 0.09 |
SMO |
smoothened, frizzled family receptor |
122 |
0.95 |
| chr8_102300419_102300649 | 0.09 |
ENSG00000239115 |
. |
19061 |
0.24 |
| chr11_70244216_70245100 | 0.09 |
CTTN |
cortactin |
11 |
0.53 |
| chr11_8788560_8788716 | 0.09 |
ST5 |
suppression of tumorigenicity 5 |
379 |
0.79 |
| chr2_232545912_232546437 | 0.09 |
MGC4771 |
|
25447 |
0.14 |
| chr20_62526165_62526370 | 0.09 |
DNAJC5 |
DnaJ (Hsp40) homolog, subfamily C, member 5 |
251 |
0.8 |
| chr12_49392080_49393040 | 0.09 |
RP11-386G11.5 |
|
379 |
0.49 |
| chr2_149894818_149895436 | 0.08 |
LYPD6B |
LY6/PLAUR domain containing 6B |
105 |
0.98 |
| chr1_232725835_232725986 | 0.08 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
28606 |
0.25 |
| chr5_16935133_16935934 | 0.08 |
MYO10 |
myosin X |
514 |
0.81 |
| chr18_6729547_6730123 | 0.08 |
ARHGAP28 |
Rho GTPase activating protein 28 |
14 |
0.5 |
| chr3_181413371_181413945 | 0.08 |
SOX2-OT |
SOX2 overlapping transcript (non-protein coding) |
3623 |
0.29 |
| chr3_50232089_50232320 | 0.08 |
GNAT1 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
2099 |
0.19 |
| chr9_110228052_110228707 | 0.08 |
ENSG00000199905 |
. |
9041 |
0.21 |
| chr10_19777738_19777947 | 0.08 |
MALRD1 |
MAM and LDL receptor class A domain containing 1 |
181 |
0.97 |
| chr5_134824456_134824665 | 0.08 |
TIFAB |
TRAF-interacting protein with forkhead-associated domain, family member B |
36471 |
0.11 |
| chr10_123914231_123914382 | 0.08 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
8635 |
0.28 |
| chr5_176924144_176924598 | 0.08 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
191 |
0.88 |
| chr11_67070981_67071360 | 0.08 |
SSH3 |
slingshot protein phosphatase 3 |
73 |
0.95 |
| chr5_177894971_177895186 | 0.08 |
CTB-26E19.1 |
|
24983 |
0.2 |
| chr14_103673729_103674048 | 0.08 |
ENSG00000239117 |
. |
4365 |
0.24 |
| chr6_135592009_135592160 | 0.08 |
RP1-32B1.4 |
|
10950 |
0.18 |
| chr6_157610005_157610158 | 0.08 |
ENSG00000252609 |
. |
102350 |
0.07 |
| chr10_110225810_110226152 | 0.08 |
ENSG00000222436 |
. |
475087 |
0.01 |
| chr22_35748941_35749092 | 0.08 |
ENSG00000266320 |
. |
17383 |
0.15 |
| chr5_78985270_78986298 | 0.08 |
CMYA5 |
cardiomyopathy associated 5 |
84 |
0.98 |
| chr2_74881397_74882034 | 0.08 |
SEMA4F |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
277 |
0.89 |
| chr16_78540334_78540585 | 0.08 |
RP11-264L1.4 |
|
6 |
0.99 |
| chr18_25758066_25758464 | 0.08 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
855 |
0.77 |
| chr5_101632211_101632449 | 0.08 |
SLCO4C1 |
solute carrier organic anion transporter family, member 4C1 |
77 |
0.98 |
| chr19_4473895_4474732 | 0.08 |
HDGFRP2 |
Hepatoma-derived growth factor-related protein 2 |
576 |
0.53 |
| chr10_74544262_74544615 | 0.08 |
RP11-354E23.5 |
|
18120 |
0.19 |
| chr22_42835920_42836170 | 0.08 |
NFAM1 |
NFAT activating protein with ITAM motif 1 |
7644 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.1 | 0.2 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.0 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.0 | GO:0005606 | laminin-1 complex(GO:0005606) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |