Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ELF3_EHF
Z-value: 1.19
Transcription factors associated with ELF3_EHF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
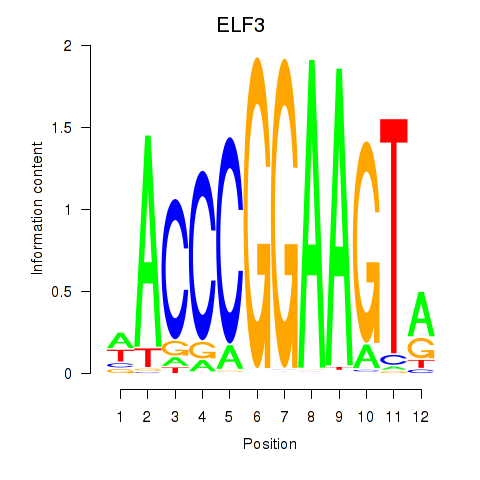
ELF3
|
ENSG00000163435.11 | E74 like ETS transcription factor 3 |
|
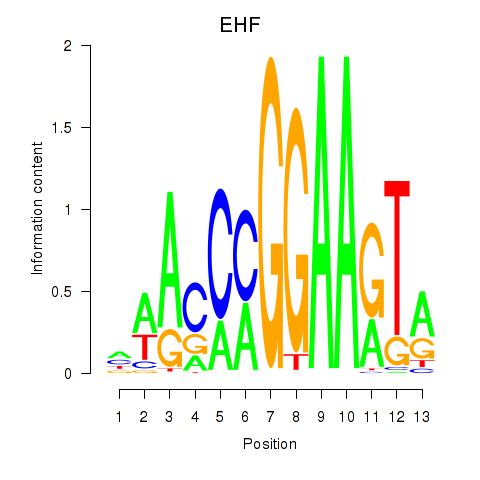
EHF
|
ENSG00000135373.8 | ETS homologous factor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_34653922_34654073 | EHF | 14 | 0.987265 | 0.81 | 8.6e-03 | Click! |
| chr11_34666501_34666652 | EHF | 2402 | 0.412860 | 0.72 | 3.0e-02 | Click! |
| chr11_34654877_34655028 | EHF | 941 | 0.698974 | 0.64 | 6.5e-02 | Click! |
| chr11_34646975_34647126 | EHF | 1227 | 0.603199 | 0.62 | 7.6e-02 | Click! |
| chr11_34653140_34653291 | EHF | 796 | 0.753427 | 0.60 | 8.5e-02 | Click! |
| chr1_201977190_201977341 | ELF3 | 192 | 0.880064 | 0.00 | 9.9e-01 | Click! |
Activity of the ELF3_EHF motif across conditions
Conditions sorted by the z-value of the ELF3_EHF motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
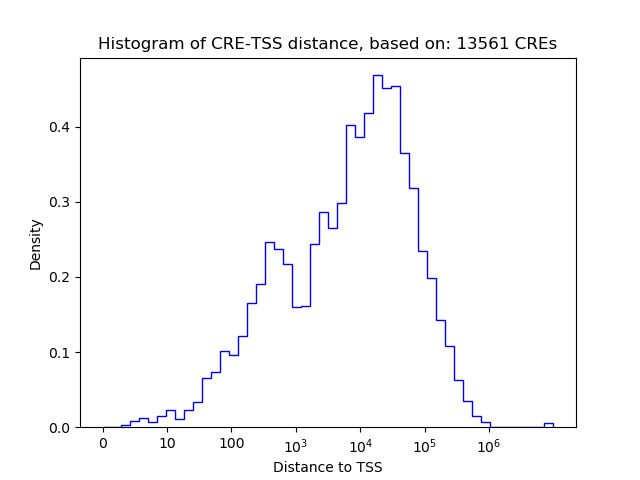
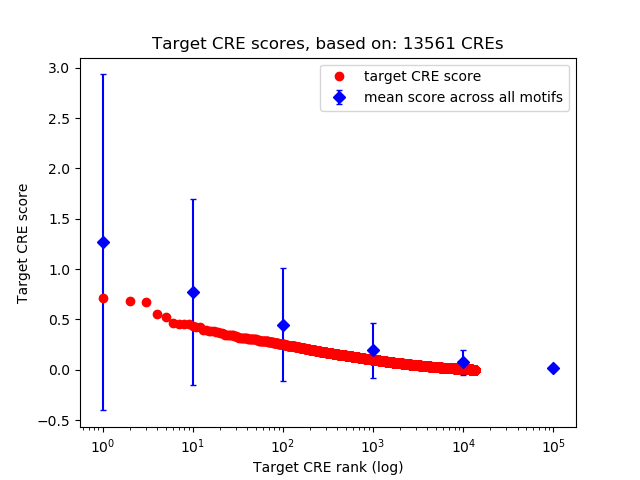
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_70244216_70245100 | 0.71 |
CTTN |
cortactin |
11 |
0.53 |
| chr13_114103770_114104137 | 0.68 |
ADPRHL1 |
ADP-ribosylhydrolase like 1 |
361 |
0.83 |
| chr16_77224906_77225760 | 0.68 |
MON1B |
MON1 secretory trafficking family member B |
236 |
0.93 |
| chr13_114103239_114103495 | 0.56 |
ADPRHL1 |
ADP-ribosylhydrolase like 1 |
90 |
0.96 |
| chr2_161826657_161826810 | 0.52 |
ENSG00000244372 |
. |
92675 |
0.09 |
| chr8_49622141_49622390 | 0.47 |
EFCAB1 |
EF-hand calcium binding domain 1 |
20083 |
0.28 |
| chr2_241949003_241950045 | 0.46 |
AC093585.6 |
|
637 |
0.62 |
| chr10_15138786_15140121 | 0.45 |
RPP38 |
ribonuclease P/MRP 38kDa subunit |
34 |
0.62 |
| chr13_30251628_30251779 | 0.45 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
81878 |
0.11 |
| chr6_116382044_116382245 | 0.43 |
FRK |
fyn-related kinase |
223 |
0.94 |
| chr22_19165246_19166141 | 0.43 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
341 |
0.82 |
| chr11_7599067_7599755 | 0.42 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
1140 |
0.49 |
| chr4_177930767_177930918 | 0.40 |
ENSG00000222859 |
. |
171754 |
0.03 |
| chr6_137555100_137555380 | 0.39 |
IFNGR1 |
interferon gamma receptor 1 |
14654 |
0.26 |
| chr13_51483605_51485004 | 0.39 |
RNASEH2B |
ribonuclease H2, subunit B |
412 |
0.48 |
| chr18_56604719_56604870 | 0.39 |
ENSG00000251870 |
. |
11266 |
0.21 |
| chr5_150273550_150273701 | 0.38 |
ZNF300 |
zinc finger protein 300 |
10726 |
0.19 |
| chr10_118764443_118765166 | 0.38 |
KIAA1598 |
KIAA1598 |
72 |
0.98 |
| chr20_3189609_3190930 | 0.37 |
ITPA |
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
98 |
0.95 |
| chr1_224712859_224713133 | 0.37 |
WDR26 |
WD repeat domain 26 |
88261 |
0.07 |
| chr13_67804057_67805835 | 0.36 |
PCDH9 |
protocadherin 9 |
478 |
0.9 |
| chr22_30831807_30831958 | 0.36 |
MTFP1 |
mitochondrial fission process 1 |
10077 |
0.07 |
| chr10_15411519_15412658 | 0.35 |
FAM171A1 |
family with sequence similarity 171, member A1 |
970 |
0.7 |
| chr11_118794725_118796211 | 0.35 |
BCL9L |
B-cell CLL/lymphoma 9-like |
5855 |
0.09 |
| chr1_214397865_214398571 | 0.35 |
SMYD2 |
SET and MYND domain containing 2 |
56358 |
0.16 |
| chr17_48714816_48714967 | 0.35 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
2673 |
0.17 |
| chr5_140562606_140562757 | 0.34 |
PCDHB16 |
protocadherin beta 16 |
1701 |
0.15 |
| chr5_54455785_54456014 | 0.34 |
GPX8 |
glutathione peroxidase 8 (putative) |
47 |
0.96 |
| chr13_26312101_26312591 | 0.33 |
AL138815.1 |
Uncharacterized protein |
129715 |
0.05 |
| chr2_228683348_228683499 | 0.33 |
CCL20 |
chemokine (C-C motif) ligand 20 |
4853 |
0.24 |
| chr10_71988541_71988692 | 0.32 |
PPA1 |
pyrophosphatase (inorganic) 1 |
4544 |
0.24 |
| chr2_128492018_128492224 | 0.32 |
SFT2D3 |
SFT2 domain containing 3 |
33524 |
0.13 |
| chr10_10836310_10836461 | 0.32 |
CELF2 |
CUGBP, Elav-like family member 2 |
210874 |
0.02 |
| chr6_1960344_1960495 | 0.32 |
GMDS |
GDP-mannose 4,6-dehydratase |
215806 |
0.02 |
| chr5_167793292_167793676 | 0.32 |
WWC1 |
WW and C2 domain containing 1 |
4942 |
0.3 |
| chr12_80747054_80747205 | 0.32 |
OTOGL |
otogelin-like |
3005 |
0.29 |
| chr2_43986075_43986259 | 0.32 |
ENSG00000252490 |
. |
13322 |
0.19 |
| chr7_150779695_150780926 | 0.32 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
9 |
0.94 |
| chr14_55571041_55571192 | 0.31 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
19712 |
0.19 |
| chr7_128021355_128021506 | 0.31 |
PRRT4 |
proline-rich transmembrane protein 4 |
19691 |
0.12 |
| chr3_105848938_105849089 | 0.31 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
260617 |
0.02 |
| chr3_126725218_126725369 | 0.31 |
PLXNA1 |
plexin A1 |
17856 |
0.27 |
| chr4_19741105_19741415 | 0.31 |
SLIT2 |
slit homolog 2 (Drosophila) |
513623 |
0.0 |
| chr6_170604225_170605407 | 0.31 |
FAM120B |
family with sequence similarity 120B |
407 |
0.83 |
| chr12_1905695_1906675 | 0.30 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
14701 |
0.2 |
| chr2_26367902_26368120 | 0.30 |
GAREML |
GRB2 associated, regulator of MAPK1-like |
27949 |
0.17 |
| chr2_23627518_23627754 | 0.30 |
KLHL29 |
kelch-like family member 29 |
19548 |
0.28 |
| chr9_96218955_96219106 | 0.30 |
FAM120AOS |
family with sequence similarity 120A opposite strand |
3156 |
0.18 |
| chr21_15922641_15922820 | 0.30 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
4066 |
0.28 |
| chr16_578205_579384 | 0.30 |
CAPN15 |
calpain 15 |
938 |
0.34 |
| chr6_117587237_117587388 | 0.30 |
VGLL2 |
vestigial like 2 (Drosophila) |
575 |
0.77 |
| chr20_1892396_1892547 | 0.30 |
SIRPA |
signal-regulatory protein alpha |
16529 |
0.21 |
| chr22_46476344_46476543 | 0.30 |
FLJ27365 |
hsa-mir-4763 |
251 |
0.85 |
| chr6_83073574_83074143 | 0.29 |
TPBG |
trophoblast glycoprotein |
103 |
0.98 |
| chr2_83156543_83156694 | 0.29 |
ENSG00000201311 |
. |
622326 |
0.0 |
| chr2_53795780_53795969 | 0.29 |
ENSG00000207456 |
. |
1743 |
0.47 |
| chr8_97172057_97172969 | 0.29 |
GDF6 |
growth differentiation factor 6 |
507 |
0.84 |
| chr3_22187235_22187386 | 0.29 |
ENSG00000221384 |
. |
316080 |
0.01 |
| chr14_50505887_50506056 | 0.29 |
ENSG00000201358 |
. |
29315 |
0.13 |
| chr5_109632882_109633033 | 0.29 |
ENSG00000221436 |
. |
216659 |
0.02 |
| chr17_62705322_62705475 | 0.29 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
47212 |
0.12 |
| chr6_1798117_1798268 | 0.28 |
FOXC1 |
forkhead box C1 |
187511 |
0.03 |
| chr12_53693277_53694636 | 0.28 |
C12orf10 |
chromosome 12 open reading frame 10 |
79 |
0.58 |
| chr2_28817013_28817303 | 0.28 |
PLB1 |
phospholipase B1 |
7628 |
0.24 |
| chr2_203881741_203882170 | 0.28 |
NBEAL1 |
neurobeachin-like 1 |
2353 |
0.35 |
| chr4_84772402_84772553 | 0.28 |
ENSG00000201633 |
. |
215034 |
0.02 |
| chr6_136417248_136417399 | 0.28 |
RP13-143G15.3 |
|
23358 |
0.2 |
| chr6_150246697_150247451 | 0.28 |
RAET1G |
retinoic acid early transcript 1G |
2817 |
0.17 |
| chr12_51640112_51640647 | 0.28 |
DAZAP2 |
DAZ associated protein 2 |
7300 |
0.14 |
| chr10_92616661_92616812 | 0.28 |
HTR7 |
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
719 |
0.67 |
| chr3_13869046_13869197 | 0.28 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
52497 |
0.13 |
| chr15_35283823_35284080 | 0.28 |
ZNF770 |
zinc finger protein 770 |
3463 |
0.25 |
| chr11_61447475_61447708 | 0.28 |
DAGLA |
diacylglycerol lipase, alpha |
314 |
0.89 |
| chr14_103389080_103389600 | 0.27 |
AMN |
amnion associated transmembrane protein |
347 |
0.84 |
| chr18_37368450_37368601 | 0.27 |
ENSG00000212354 |
. |
109939 |
0.07 |
| chr10_18126007_18126187 | 0.27 |
ENSG00000207938 |
. |
7939 |
0.2 |
| chr2_217204685_217204878 | 0.27 |
ENSG00000207303 |
. |
11345 |
0.17 |
| chr15_71184312_71185443 | 0.27 |
LRRC49 |
leucine rich repeat containing 49 |
88 |
0.55 |
| chr19_55862099_55862250 | 0.27 |
COX6B2 |
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
3734 |
0.08 |
| chr5_106900739_106900890 | 0.27 |
EFNA5 |
ephrin-A5 |
105514 |
0.08 |
| chr6_37227035_37227379 | 0.27 |
TBC1D22B |
TBC1 domain family, member 22B |
1659 |
0.28 |
| chr14_35400431_35400683 | 0.27 |
ENSG00000199980 |
. |
2192 |
0.28 |
| chr1_11850709_11850962 | 0.27 |
C1orf167 |
chromosome 1 open reading frame 167 |
10961 |
0.11 |
| chr8_145925356_145925968 | 0.26 |
AF186192.5 |
|
5257 |
0.15 |
| chr17_15870184_15870534 | 0.26 |
ADORA2B |
adenosine A2b receptor |
22128 |
0.14 |
| chr17_38255735_38256962 | 0.26 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
630 |
0.6 |
| chr2_135105545_135105696 | 0.26 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
93790 |
0.09 |
| chr10_69865396_69865547 | 0.26 |
MYPN |
myopalladin |
441 |
0.84 |
| chr10_375832_375983 | 0.26 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
441 |
0.85 |
| chr5_52367762_52367913 | 0.26 |
CTD-2366F13.2 |
|
17461 |
0.19 |
| chr2_28672824_28673107 | 0.26 |
PLB1 |
phospholipase B1 |
7047 |
0.22 |
| chr2_23603831_23604295 | 0.26 |
KLHL29 |
kelch-like family member 29 |
4025 |
0.36 |
| chr13_107137023_107137174 | 0.26 |
EFNB2 |
ephrin-B2 |
50364 |
0.17 |
| chr5_176074525_176075535 | 0.26 |
TSPAN17 |
tetraspanin 17 |
409 |
0.77 |
| chr17_2612430_2612749 | 0.26 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
1903 |
0.21 |
| chr1_33207592_33208218 | 0.25 |
KIAA1522 |
KIAA1522 |
419 |
0.8 |
| chr3_9446443_9446669 | 0.25 |
SETD5-AS1 |
SETD5 antisense RNA 1 |
6293 |
0.15 |
| chr18_31738936_31739121 | 0.25 |
NOL4 |
nucleolar protein 4 |
63028 |
0.13 |
| chr19_55791240_55791709 | 0.25 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
5 |
0.94 |
| chr2_18093683_18093834 | 0.25 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
33108 |
0.21 |
| chr9_34663128_34663279 | 0.25 |
CCL27 |
chemokine (C-C motif) ligand 27 |
514 |
0.56 |
| chr7_17720120_17720505 | 0.25 |
SNX13 |
sorting nexin 13 |
259779 |
0.02 |
| chr10_33622786_33623487 | 0.25 |
NRP1 |
neuropilin 1 |
174 |
0.97 |
| chr10_12376015_12376474 | 0.25 |
ENSG00000264036 |
. |
5856 |
0.19 |
| chr4_86419636_86419834 | 0.25 |
ARHGAP24 |
Rho GTPase activating protein 24 |
23344 |
0.28 |
| chr3_184504176_184504876 | 0.25 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
25405 |
0.22 |
| chr3_189348802_189349006 | 0.24 |
TP63 |
tumor protein p63 |
312 |
0.95 |
| chr9_97785492_97785727 | 0.24 |
C9orf3 |
chromosome 9 open reading frame 3 |
18331 |
0.17 |
| chr1_27884100_27884415 | 0.24 |
RP1-159A19.4 |
|
31941 |
0.12 |
| chr17_39673114_39673265 | 0.24 |
KRT15 |
keratin 15 |
1527 |
0.2 |
| chr4_184922100_184922251 | 0.24 |
STOX2 |
storkhead box 2 |
303 |
0.92 |
| chr3_54121860_54122011 | 0.24 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
34639 |
0.22 |
| chr15_71373754_71373905 | 0.24 |
THSD4 |
thrombospondin, type I, domain containing 4 |
15462 |
0.22 |
| chrX_23815798_23815949 | 0.24 |
SAT1 |
spermidine/spermine N1-acetyltransferase 1 |
14583 |
0.14 |
| chr9_25677269_25678110 | 0.24 |
TUSC1 |
tumor suppressor candidate 1 |
1167 |
0.68 |
| chr7_100660155_100661047 | 0.24 |
RP11-395B7.4 |
|
281 |
0.85 |
| chr11_27485860_27486011 | 0.24 |
RP11-159H22.2 |
|
7341 |
0.17 |
| chr6_106994445_106994613 | 0.24 |
AIM1 |
absent in melanoma 1 |
5500 |
0.23 |
| chr14_103394355_103395516 | 0.24 |
AMN |
amnion associated transmembrane protein |
211 |
0.92 |
| chr11_44338057_44338887 | 0.24 |
ALX4 |
ALX homeobox 4 |
6756 |
0.31 |
| chr3_50194015_50194506 | 0.24 |
RP11-493K19.3 |
|
742 |
0.47 |
| chr1_44513271_44513499 | 0.24 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
16251 |
0.14 |
| chr4_81189966_81190509 | 0.24 |
FGF5 |
fibroblast growth factor 5 |
2444 |
0.36 |
| chr3_113616260_113616494 | 0.24 |
GRAMD1C |
GRAM domain containing 1C |
38 |
0.98 |
| chr2_103405948_103406099 | 0.24 |
TMEM182 |
transmembrane protein 182 |
27551 |
0.22 |
| chr2_64821582_64821733 | 0.24 |
ENSG00000252414 |
. |
15631 |
0.2 |
| chr22_38452981_38453959 | 0.24 |
PICK1 |
protein interacting with PRKCA 1 |
56 |
0.95 |
| chr5_167646669_167646982 | 0.24 |
CTB-178M22.2 |
|
12537 |
0.22 |
| chr18_21407330_21407481 | 0.23 |
LAMA3 |
laminin, alpha 3 |
6512 |
0.26 |
| chr12_6798179_6798744 | 0.23 |
ZNF384 |
zinc finger protein 384 |
2 |
0.94 |
| chr19_2014732_2015442 | 0.23 |
BTBD2 |
BTB (POZ) domain containing 2 |
632 |
0.54 |
| chr5_58817449_58817600 | 0.23 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
64695 |
0.15 |
| chr6_12314255_12314578 | 0.23 |
EDN1 |
endothelin 1 |
23820 |
0.26 |
| chr6_27263961_27264170 | 0.23 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
15026 |
0.2 |
| chr14_20928526_20929515 | 0.23 |
TMEM55B |
transmembrane protein 55B |
32 |
0.94 |
| chr11_8829373_8829660 | 0.23 |
ST5 |
suppression of tumorigenicity 5 |
2680 |
0.22 |
| chr19_12780320_12781072 | 0.23 |
WDR83 |
WD repeat domain 83 |
179 |
0.62 |
| chr3_134261428_134261579 | 0.23 |
CEP63 |
centrosomal protein 63kDa |
3495 |
0.28 |
| chr1_147148877_147149210 | 0.23 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
6425 |
0.22 |
| chr2_196444875_196445026 | 0.23 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
4249 |
0.31 |
| chr20_32856950_32857430 | 0.23 |
ASIP |
agouti signaling protein |
9019 |
0.15 |
| chrX_129473317_129474471 | 0.23 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
7 |
0.98 |
| chrX_54947220_54948504 | 0.23 |
TRO |
trophinin |
41 |
0.97 |
| chrX_129838124_129838315 | 0.23 |
ENOX2 |
ecto-NOX disulfide-thiol exchanger 2 |
5068 |
0.34 |
| chr9_112674232_112674383 | 0.22 |
PALM2 |
paralemmin 2 |
44996 |
0.18 |
| chr20_1874240_1874814 | 0.22 |
SIRPA |
signal-regulatory protein alpha |
627 |
0.78 |
| chr1_2187001_2187152 | 0.22 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
26942 |
0.11 |
| chr7_130602447_130602598 | 0.22 |
ENSG00000226380 |
. |
40224 |
0.17 |
| chr10_95172691_95172842 | 0.22 |
MYOF |
myoferlin |
69185 |
0.1 |
| chr20_37360527_37360678 | 0.22 |
SLC32A1 |
solute carrier family 32 (GABA vesicular transporter), member 1 |
7497 |
0.17 |
| chr4_77905705_77905940 | 0.22 |
SEPT11 |
septin 11 |
355 |
0.91 |
| chr3_169755608_169756727 | 0.22 |
GPR160 |
G protein-coupled receptor 160 |
241 |
0.91 |
| chr7_98970111_98970529 | 0.22 |
ARPC1B |
actin related protein 2/3 complex, subunit 1B, 41kDa |
1552 |
0.28 |
| chr6_105388737_105389249 | 0.22 |
LIN28B |
lin-28 homolog B (C. elegans) |
15930 |
0.23 |
| chr4_38611399_38611550 | 0.22 |
RP11-617D20.1 |
|
14722 |
0.22 |
| chr20_18296318_18296469 | 0.22 |
RP4-568F9.3 |
|
638 |
0.6 |
| chr1_84831627_84831778 | 0.22 |
SAMD13 |
sterile alpha motif domain containing 13 |
21183 |
0.19 |
| chr12_53264228_53264379 | 0.22 |
KRT78 |
keratin 78 |
21427 |
0.12 |
| chr10_18446622_18446802 | 0.22 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
16603 |
0.26 |
| chr3_15332172_15332523 | 0.22 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
14795 |
0.14 |
| chr5_142185385_142185536 | 0.22 |
ARHGAP26 |
Rho GTPase activating protein 26 |
33178 |
0.2 |
| chr2_145765254_145765507 | 0.22 |
ENSG00000253036 |
. |
327258 |
0.01 |
| chr13_99645097_99645248 | 0.22 |
DOCK9 |
dedicator of cytokinesis 9 |
14834 |
0.22 |
| chr4_10007868_10008019 | 0.22 |
RP11-448G15.1 |
|
163 |
0.95 |
| chr4_72125821_72125972 | 0.22 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
72852 |
0.13 |
| chr2_125926045_125926196 | 0.22 |
ENSG00000201853 |
. |
39711 |
0.23 |
| chr8_135433860_135434011 | 0.21 |
ZFAT |
zinc finger and AT hook domain containing |
88490 |
0.1 |
| chr15_33121037_33121231 | 0.21 |
FMN1 |
formin 1 |
59321 |
0.11 |
| chr17_77767116_77767731 | 0.21 |
CBX8 |
chromobox homolog 8 |
3492 |
0.16 |
| chr5_443562_444067 | 0.21 |
EXOC3 |
exocyst complex component 3 |
431 |
0.52 |
| chr2_151339584_151339865 | 0.21 |
RND3 |
Rho family GTPase 3 |
2172 |
0.49 |
| chr1_1676260_1677404 | 0.21 |
SLC35E2 |
solute carrier family 35, member E2 |
526 |
0.68 |
| chr10_7306332_7306626 | 0.21 |
SFMBT2 |
Scm-like with four mbt domains 2 |
144228 |
0.05 |
| chr8_107965641_107965792 | 0.21 |
ABRA |
actin-binding Rho activating protein |
183243 |
0.03 |
| chr1_91189429_91189597 | 0.21 |
BARHL2 |
BarH-like homeobox 2 |
6719 |
0.31 |
| chr1_201228727_201229116 | 0.21 |
PKP1 |
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
23659 |
0.15 |
| chrX_69674453_69675874 | 0.21 |
DLG3 |
discs, large homolog 3 (Drosophila) |
217 |
0.75 |
| chr1_65430726_65432146 | 0.21 |
JAK1 |
Janus kinase 1 |
751 |
0.73 |
| chr5_139927168_139928105 | 0.21 |
EIF4EBP3 |
eukaryotic translation initiation factor 4E binding protein 3 |
385 |
0.69 |
| chr6_160015485_160015820 | 0.21 |
SOD2 |
superoxide dismutase 2, mitochondrial |
98608 |
0.07 |
| chr19_13906290_13907522 | 0.21 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
632 |
0.39 |
| chr2_208095383_208095534 | 0.21 |
AC007879.5 |
|
23518 |
0.19 |
| chr9_78796062_78796213 | 0.21 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
7418 |
0.32 |
| chr1_173854808_173854959 | 0.21 |
ZBTB37 |
zinc finger and BTB domain containing 37 |
16789 |
0.1 |
| chr4_54174706_54174935 | 0.21 |
ENSG00000207385 |
. |
43086 |
0.16 |
| chr19_56166514_56167932 | 0.21 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
792 |
0.34 |
| chr7_41223414_41223565 | 0.21 |
AC005160.3 |
|
408332 |
0.01 |
| chr16_4710556_4710707 | 0.21 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
10230 |
0.11 |
| chr2_242495139_242495290 | 0.21 |
BOK |
BCL2-related ovarian killer |
2922 |
0.17 |
| chr10_99608959_99609709 | 0.21 |
GOLGA7B |
golgin A7 family, member B |
662 |
0.7 |
| chr5_158689547_158690875 | 0.20 |
UBLCP1 |
ubiquitin-like domain containing CTD phosphatase 1 |
122 |
0.96 |
| chr2_102359188_102359339 | 0.20 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
44271 |
0.19 |
| chr2_176931780_176932252 | 0.20 |
EVX2 |
even-skipped homeobox 2 |
16625 |
0.1 |
| chr21_35348616_35348984 | 0.20 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
6980 |
0.21 |
| chr16_75046673_75046884 | 0.20 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
10583 |
0.2 |
| chr10_63642613_63642764 | 0.20 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
18371 |
0.24 |
| chr8_49835583_49835808 | 0.20 |
SNAI2 |
snail family zinc finger 2 |
1396 |
0.59 |
| chr9_117868310_117868461 | 0.20 |
TNC |
tenascin C |
6651 |
0.27 |
| chr2_201171688_201172904 | 0.20 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
701 |
0.76 |
| chr6_85472920_85473300 | 0.20 |
TBX18 |
T-box 18 |
37 |
0.99 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.1 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.1 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.2 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.2 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.1 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.2 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.2 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.2 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0060856 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0072077 | renal vesicle morphogenesis(GO:0072077) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.2 | GO:0046040 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0031622 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.0 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.0 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.2 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) |
| 0.0 | 0.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.0 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:1902692 | regulation of neuroblast proliferation(GO:1902692) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:0032276 | regulation of gonadotropin secretion(GO:0032276) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.0 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:0031128 | organ induction(GO:0001759) developmental induction(GO:0031128) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0061515 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.0 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.2 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.1 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 1.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |