Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
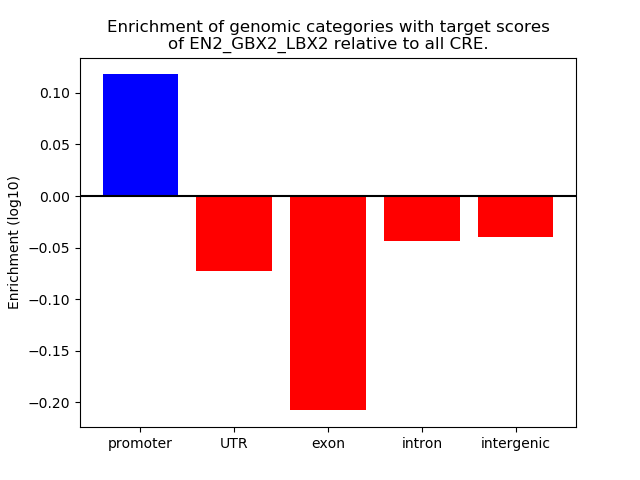
Results for EN2_GBX2_LBX2
Z-value: 0.30
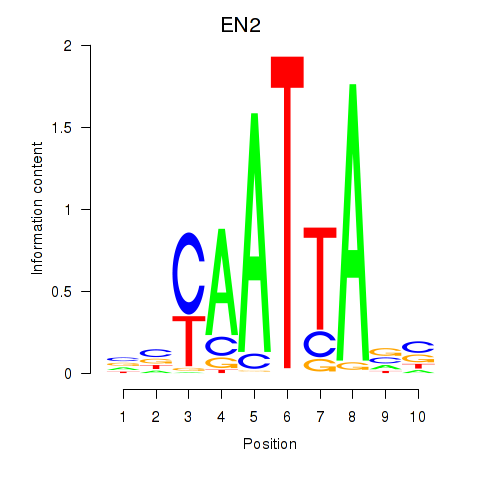
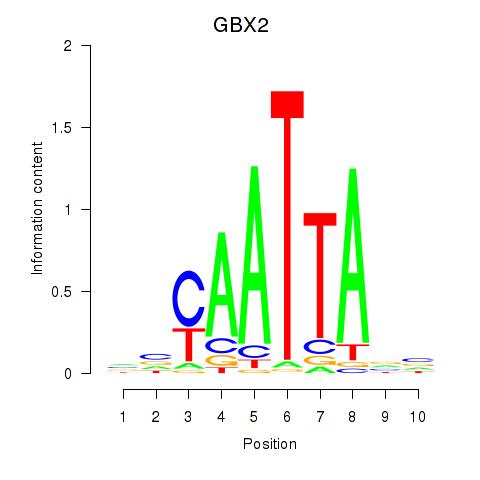
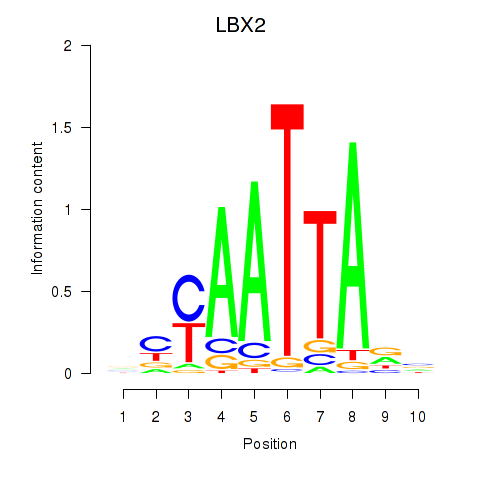
Transcription factors associated with EN2_GBX2_LBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EN2
|
ENSG00000164778.4 | engrailed homeobox 2 |
|
GBX2
|
ENSG00000168505.6 | gastrulation brain homeobox 2 |
|
LBX2
|
ENSG00000179528.11 | ladybird homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_155259792_155259943 | EN2 | 9043 | 0.196903 | 0.48 | 1.9e-01 | Click! |
| chr7_155260838_155261005 | EN2 | 10097 | 0.193888 | 0.39 | 3.0e-01 | Click! |
| chr7_155250668_155250957 | EN2 | 12 | 0.979457 | -0.31 | 4.1e-01 | Click! |
| chr7_155265207_155265358 | EN2 | 14458 | 0.183389 | -0.28 | 4.6e-01 | Click! |
| chr7_155261127_155261495 | EN2 | 10487 | 0.192858 | 0.28 | 4.7e-01 | Click! |
| chr2_237076524_237076816 | GBX2 | 18 | 0.912238 | 0.48 | 1.9e-01 | Click! |
| chr2_237081096_237081247 | GBX2 | 4159 | 0.174508 | 0.44 | 2.4e-01 | Click! |
| chr2_237077720_237077871 | GBX2 | 783 | 0.524265 | -0.35 | 3.6e-01 | Click! |
| chr2_237080254_237080405 | GBX2 | 3317 | 0.189489 | -0.34 | 3.7e-01 | Click! |
| chr2_237088721_237088872 | GBX2 | 11784 | 0.144978 | 0.31 | 4.1e-01 | Click! |
| chr2_74730587_74730945 | LBX2 | 323 | 0.619077 | 0.61 | 8.1e-02 | Click! |
| chr2_74729754_74730571 | LBX2 | 51 | 0.816052 | 0.44 | 2.3e-01 | Click! |
| chr2_74726789_74726940 | LBX2 | 21 | 0.808069 | 0.41 | 2.7e-01 | Click! |
| chr2_74732376_74732527 | LBX2 | 2008 | 0.103893 | 0.40 | 2.9e-01 | Click! |
| chr2_74731433_74731584 | LBX2 | 1065 | 0.203023 | 0.10 | 8.0e-01 | Click! |
Activity of the EN2_GBX2_LBX2 motif across conditions
Conditions sorted by the z-value of the EN2_GBX2_LBX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
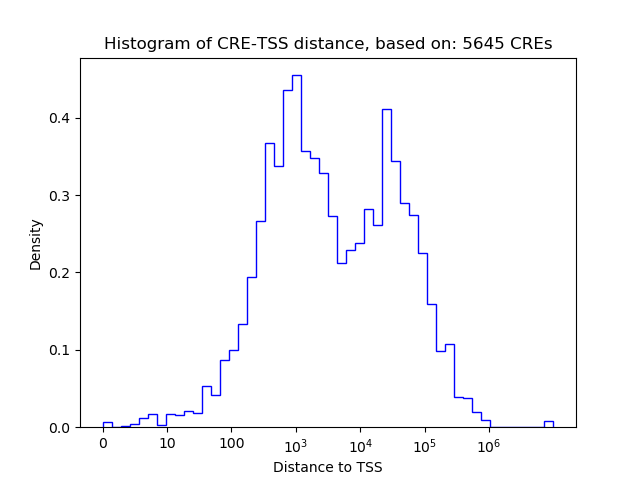
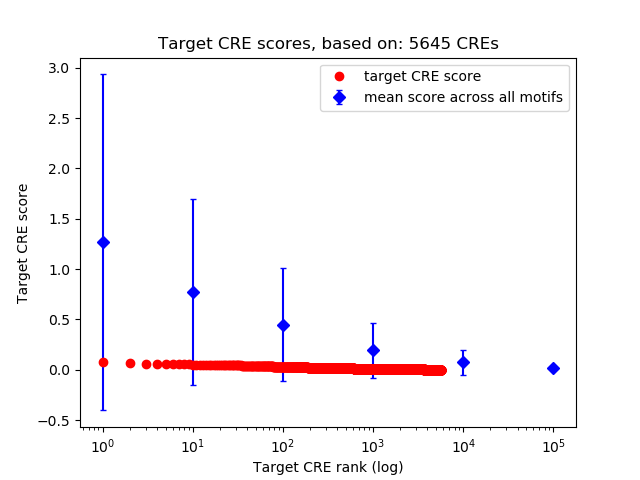
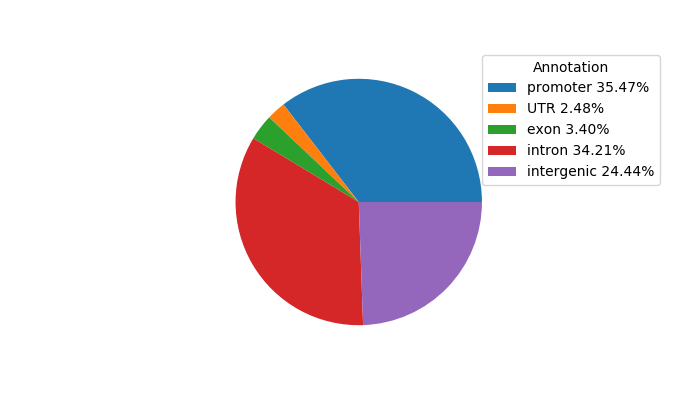
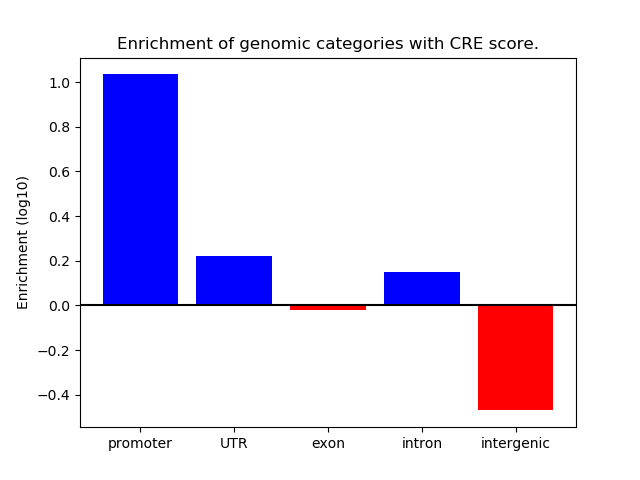
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_94661378_94661529 | 0.07 |
RP11-856F16.2 |
|
2767 |
0.3 |
| chr10_116584415_116584656 | 0.07 |
FAM160B1 |
family with sequence similarity 160, member B1 |
3006 |
0.37 |
| chr4_144312492_144312643 | 0.06 |
GAB1 |
GRB2-associated binding protein 1 |
96 |
0.98 |
| chr6_681837_682046 | 0.06 |
EXOC2 |
exocyst complex component 2 |
11170 |
0.25 |
| chr13_45242068_45242279 | 0.06 |
ENSG00000238932 |
. |
39364 |
0.2 |
| chr13_97875674_97876020 | 0.06 |
MBNL2 |
muscleblind-like splicing regulator 2 |
1238 |
0.62 |
| chr1_28834546_28835031 | 0.06 |
ENSG00000200087 |
. |
283 |
0.74 |
| chr5_102897969_102898137 | 0.06 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
437 |
0.91 |
| chr14_103058041_103058305 | 0.05 |
RCOR1 |
REST corepressor 1 |
825 |
0.6 |
| chr4_109532705_109532935 | 0.05 |
RPL34 |
ribosomal protein L34 |
8902 |
0.2 |
| chr13_96295623_96295774 | 0.05 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
181 |
0.96 |
| chr10_91461400_91461679 | 0.05 |
KIF20B |
kinesin family member 20B |
103 |
0.98 |
| chr3_79067433_79067887 | 0.05 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
34 |
0.99 |
| chr14_76604893_76605206 | 0.05 |
GPATCH2L |
G patch domain containing 2-like |
13213 |
0.26 |
| chr3_112356169_112356393 | 0.05 |
CCDC80 |
coiled-coil domain containing 80 |
663 |
0.77 |
| chr5_134370433_134370584 | 0.05 |
PITX1 |
paired-like homeodomain 1 |
5 |
0.97 |
| chr5_77822199_77822489 | 0.05 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
22630 |
0.27 |
| chr1_238027813_238027997 | 0.05 |
RP11-193H5.1 |
|
2430 |
0.36 |
| chr11_27406666_27406921 | 0.05 |
CCDC34 |
coiled-coil domain containing 34 |
21378 |
0.22 |
| chr10_105211471_105211777 | 0.05 |
CALHM2 |
calcium homeostasis modulator 2 |
451 |
0.57 |
| chr9_133711223_133711921 | 0.05 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
1119 |
0.56 |
| chr15_50648671_50648965 | 0.05 |
ENSG00000244879 |
. |
150 |
0.91 |
| chr11_61158553_61158704 | 0.05 |
TMEM216 |
transmembrane protein 216 |
531 |
0.65 |
| chr11_65324098_65324348 | 0.05 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
1013 |
0.31 |
| chr1_115258427_115259019 | 0.05 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
792 |
0.54 |
| chr19_2962281_2962480 | 0.05 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
15064 |
0.12 |
| chr1_16677722_16678052 | 0.05 |
FBXO42 |
F-box protein 42 |
1062 |
0.47 |
| chr10_70230757_70231440 | 0.05 |
DNA2 |
DNA replication helicase/nuclease 2 |
545 |
0.71 |
| chr7_6654683_6655004 | 0.05 |
ZNF853 |
zinc finger protein 853 |
405 |
0.77 |
| chr5_125758847_125759062 | 0.05 |
GRAMD3 |
GRAM domain containing 3 |
85 |
0.98 |
| chr7_18535021_18535832 | 0.04 |
HDAC9 |
histone deacetylase 9 |
47 |
0.99 |
| chr1_196577911_196578299 | 0.04 |
KCNT2 |
potassium channel, subfamily T, member 2 |
250 |
0.94 |
| chr8_87188438_87188625 | 0.04 |
CTD-3118D11.3 |
|
4116 |
0.24 |
| chr5_178956771_178956922 | 0.04 |
RUFY1 |
RUN and FYVE domain containing 1 |
20713 |
0.17 |
| chr1_101359723_101359874 | 0.04 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
422 |
0.6 |
| chr1_245461989_245462162 | 0.04 |
RP11-62I21.1 |
|
64272 |
0.12 |
| chr6_16486388_16486539 | 0.04 |
ENSG00000265642 |
. |
57709 |
0.16 |
| chr3_159483781_159484285 | 0.04 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1141 |
0.44 |
| chr11_12716213_12716424 | 0.04 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
19268 |
0.26 |
| chr1_177953163_177953341 | 0.04 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
13904 |
0.3 |
| chr18_67822225_67822376 | 0.04 |
RTTN |
rotatin |
50662 |
0.17 |
| chr9_136215868_136216170 | 0.04 |
ENSG00000206611 |
. |
232 |
0.42 |
| chr6_52440366_52440800 | 0.04 |
TRAM2 |
translocation associated membrane protein 2 |
1130 |
0.59 |
| chr12_116008040_116008436 | 0.04 |
ENSG00000240205 |
. |
147292 |
0.05 |
| chr4_89522534_89522685 | 0.04 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
4336 |
0.21 |
| chr2_101620301_101620452 | 0.04 |
RPL31 |
ribosomal protein L31 |
1175 |
0.38 |
| chr16_77619617_77619768 | 0.04 |
ENSG00000221226 |
. |
124506 |
0.05 |
| chr1_88150357_88150552 | 0.04 |
ENSG00000199318 |
. |
231398 |
0.02 |
| chr6_53928552_53928746 | 0.04 |
MLIP-AS1 |
MLIP antisense RNA 1 |
15875 |
0.2 |
| chr17_37007920_37008364 | 0.04 |
ENSG00000252699 |
. |
230 |
0.78 |
| chr18_2573280_2573731 | 0.04 |
NDC80 |
NDC80 kinetochore complex component |
1941 |
0.23 |
| chr6_26313507_26313734 | 0.04 |
HIST1H4H |
histone cluster 1, H4h |
27858 |
0.07 |
| chr3_87038506_87038677 | 0.04 |
VGLL3 |
vestigial like 3 (Drosophila) |
1261 |
0.64 |
| chr6_119027020_119027704 | 0.04 |
CEP85L |
centrosomal protein 85kDa-like |
3869 |
0.35 |
| chr2_85831784_85832049 | 0.04 |
TMEM150A |
transmembrane protein 150A |
2095 |
0.16 |
| chr2_224808584_224808735 | 0.04 |
WDFY1 |
WD repeat and FYVE domain containing 1 |
1415 |
0.42 |
| chr11_68924666_68924817 | 0.04 |
RP11-554A11.9 |
|
1363 |
0.5 |
| chr4_146467351_146467609 | 0.04 |
SMAD1-AS1 |
SMAD1 antisense RNA 1 |
29210 |
0.17 |
| chr8_22023419_22024083 | 0.04 |
BMP1 |
bone morphogenetic protein 1 |
951 |
0.4 |
| chr15_57516945_57517146 | 0.04 |
TCF12 |
transcription factor 12 |
5381 |
0.29 |
| chr2_675828_676052 | 0.04 |
TMEM18 |
transmembrane protein 18 |
165 |
0.93 |
| chr4_153511797_153511976 | 0.04 |
ENSG00000268471 |
. |
54306 |
0.12 |
| chr3_156469026_156469297 | 0.04 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
70206 |
0.1 |
| chr18_19612717_19612880 | 0.03 |
ENSG00000265568 |
. |
33268 |
0.15 |
| chr10_116474996_116475239 | 0.03 |
ABLIM1 |
actin binding LIM protein 1 |
30703 |
0.21 |
| chr12_66220556_66220804 | 0.03 |
HMGA2 |
high mobility group AT-hook 2 |
1777 |
0.39 |
| chr2_197832046_197832264 | 0.03 |
ANKRD44 |
ankyrin repeat domain 44 |
33071 |
0.18 |
| chr15_96878465_96878729 | 0.03 |
ENSG00000222651 |
. |
2107 |
0.24 |
| chr1_8087615_8088020 | 0.03 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
1449 |
0.45 |
| chr5_14039928_14040178 | 0.03 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
95401 |
0.09 |
| chr4_152337049_152337200 | 0.03 |
FAM160A1 |
family with sequence similarity 160, member A1 |
6620 |
0.28 |
| chr16_24742200_24742688 | 0.03 |
TNRC6A |
trinucleotide repeat containing 6A |
1410 |
0.55 |
| chr2_217235630_217236249 | 0.03 |
MARCH4 |
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
811 |
0.59 |
| chr3_16024121_16024347 | 0.03 |
ENSG00000207815 |
. |
108956 |
0.06 |
| chr12_133347795_133347946 | 0.03 |
ANKLE2 |
ankyrin repeat and LEM domain containing 2 |
9396 |
0.15 |
| chr15_101261674_101262051 | 0.03 |
ENSG00000212306 |
. |
83294 |
0.08 |
| chr10_52750817_52751095 | 0.03 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
11 |
0.99 |
| chr12_54120032_54120198 | 0.03 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
9 |
0.98 |
| chr22_46476561_46476775 | 0.03 |
FLJ27365 |
hsa-mir-4763 |
476 |
0.66 |
| chr8_110619590_110619741 | 0.03 |
SYBU |
syntabulin (syntaxin-interacting) |
639 |
0.72 |
| chr11_16759613_16759958 | 0.03 |
C11orf58 |
chromosome 11 open reading frame 58 |
163 |
0.97 |
| chr17_47547430_47548013 | 0.03 |
NGFR |
nerve growth factor receptor |
24934 |
0.13 |
| chr2_189158529_189158680 | 0.03 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
59 |
0.98 |
| chr18_11852846_11853378 | 0.03 |
RP11-78A19.3 |
|
362 |
0.77 |
| chr10_27129753_27129904 | 0.03 |
ABI1 |
abl-interactor 1 |
19964 |
0.17 |
| chr19_49121111_49121642 | 0.03 |
RPL18 |
ribosomal protein L18 |
235 |
0.79 |
| chr1_214723042_214723197 | 0.03 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
1447 |
0.54 |
| chr11_20179312_20179463 | 0.03 |
DBX1 |
developing brain homeobox 1 |
2483 |
0.35 |
| chr1_205342500_205342651 | 0.03 |
LEMD1-AS1 |
LEMD1 antisense RNA 1 |
195 |
0.93 |
| chrX_118612161_118612324 | 0.03 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
9879 |
0.14 |
| chr6_149256843_149257025 | 0.03 |
RP11-162J8.2 |
|
28886 |
0.23 |
| chr7_26141990_26142141 | 0.03 |
ENSG00000266430 |
. |
42106 |
0.14 |
| chr13_107160462_107160937 | 0.03 |
EFNB2 |
ephrin-B2 |
26763 |
0.24 |
| chr14_80678419_80678624 | 0.03 |
DIO2 |
deiodinase, iodothyronine, type II |
0 |
0.95 |
| chr14_29859562_29859789 | 0.03 |
ENSG00000257522 |
. |
11813 |
0.29 |
| chr22_25388088_25388316 | 0.03 |
KIAA1671 |
KIAA1671 |
35739 |
0.16 |
| chr10_53126976_53127127 | 0.03 |
RP11-539E19.2 |
|
64725 |
0.12 |
| chr6_3390423_3390626 | 0.03 |
SLC22A23 |
solute carrier family 22, member 23 |
49128 |
0.15 |
| chr9_112748695_112748951 | 0.03 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
62055 |
0.14 |
| chr2_188418561_188419245 | 0.03 |
AC007319.1 |
|
27 |
0.69 |
| chr7_107640774_107640963 | 0.03 |
ENSG00000238297 |
. |
441 |
0.76 |
| chr6_49794571_49794884 | 0.03 |
RP3-417L20.4 |
|
9584 |
0.19 |
| chr16_67513595_67513746 | 0.03 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
1312 |
0.23 |
| chr9_13278507_13279169 | 0.03 |
RP11-272P10.2 |
|
348 |
0.67 |
| chr6_85896028_85896266 | 0.03 |
NT5E |
5'-nucleotidase, ecto (CD73) |
263662 |
0.02 |
| chr1_225610206_225610357 | 0.03 |
LBR |
lamin B receptor |
5318 |
0.2 |
| chr8_145562079_145562230 | 0.03 |
SCRT1 |
scratch family zinc finger 1 |
2211 |
0.12 |
| chr12_112845942_112846255 | 0.03 |
RPL6 |
ribosomal protein L6 |
658 |
0.71 |
| chr10_102921756_102921944 | 0.03 |
RP11-31L23.3 |
|
22382 |
0.12 |
| chr19_35158241_35158444 | 0.03 |
SCGB2B2 |
secretoglobin, family 2B, member 2 |
8262 |
0.14 |
| chr6_37665761_37665951 | 0.03 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
90 |
0.98 |
| chr4_47916858_47917055 | 0.03 |
NFXL1 |
nuclear transcription factor, X-box binding-like 1 |
323 |
0.9 |
| chr18_65188257_65188408 | 0.03 |
DSEL |
dermatan sulfate epimerase-like |
4115 |
0.29 |
| chr1_89992055_89992206 | 0.03 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
1668 |
0.45 |
| chr2_55458779_55458930 | 0.03 |
RPS27A |
ribosomal protein S27a |
185 |
0.75 |
| chr1_212607133_212607429 | 0.03 |
NENF |
neudesin neurotrophic factor |
1052 |
0.52 |
| chr15_59064816_59065008 | 0.03 |
FAM63B |
family with sequence similarity 63, member B |
1351 |
0.36 |
| chr20_52749591_52749788 | 0.03 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
38803 |
0.16 |
| chr17_40574158_40574426 | 0.03 |
PTRF |
polymerase I and transcript release factor |
1243 |
0.31 |
| chr21_46629242_46629401 | 0.03 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
37764 |
0.14 |
| chr16_85202371_85202571 | 0.03 |
CTC-786C10.1 |
|
2411 |
0.35 |
| chr3_45732221_45732372 | 0.03 |
SACM1L |
SAC1 suppressor of actin mutations 1-like (yeast) |
1345 |
0.36 |
| chr7_93634348_93634499 | 0.03 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
729 |
0.72 |
| chr8_145134632_145134783 | 0.03 |
EXOSC4 |
exosome component 4 |
381 |
0.5 |
| chr9_122131014_122131286 | 0.03 |
BRINP1 |
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
553 |
0.87 |
| chr5_39074216_39074429 | 0.03 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
169 |
0.97 |
| chr6_155428404_155428796 | 0.03 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
14450 |
0.28 |
| chr1_221914094_221914536 | 0.03 |
DUSP10 |
dual specificity phosphatase 10 |
1146 |
0.66 |
| chr1_153918448_153918692 | 0.03 |
DENND4B |
DENN/MADD domain containing 4B |
602 |
0.54 |
| chr4_81198150_81198317 | 0.03 |
FGF5 |
fibroblast growth factor 5 |
10440 |
0.25 |
| chr11_131780111_131780399 | 0.03 |
NTM |
neurotrimin |
642 |
0.77 |
| chr15_50410621_50410772 | 0.03 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
723 |
0.73 |
| chr8_131271894_131272045 | 0.03 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
12681 |
0.28 |
| chr17_79010696_79011070 | 0.03 |
BAIAP2 |
BAI1-associated protein 2 |
1080 |
0.4 |
| chr12_106812256_106812407 | 0.03 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
36581 |
0.19 |
| chr2_157197921_157198176 | 0.03 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
812 |
0.72 |
| chr4_170580994_170582031 | 0.03 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
299 |
0.93 |
| chr10_6763782_6764322 | 0.03 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr1_109236074_109236443 | 0.03 |
PRPF38B |
pre-mRNA processing factor 38B |
1295 |
0.4 |
| chr12_131796680_131796831 | 0.03 |
ENSG00000212251 |
. |
12459 |
0.23 |
| chr17_18091438_18091835 | 0.03 |
RP11-258F1.1 |
|
3569 |
0.14 |
| chr5_88129391_88129549 | 0.03 |
MEF2C |
myocyte enhancer factor 2C |
6495 |
0.28 |
| chr8_94507892_94508285 | 0.03 |
ENSG00000222416 |
. |
54583 |
0.14 |
| chr10_76971114_76971464 | 0.03 |
VDAC2 |
voltage-dependent anion channel 2 |
75 |
0.97 |
| chr17_37774592_37774840 | 0.03 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
8277 |
0.12 |
| chr3_81474618_81474769 | 0.03 |
ENSG00000222389 |
. |
83934 |
0.11 |
| chr4_186732947_186733362 | 0.03 |
SORBS2 |
sorbin and SH3 domain containing 2 |
21 |
0.99 |
| chr10_5853342_5853785 | 0.03 |
GDI2 |
GDP dissociation inhibitor 2 |
1787 |
0.37 |
| chr11_12791215_12791367 | 0.03 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
24703 |
0.2 |
| chr10_106007564_106007715 | 0.03 |
GSTO1 |
glutathione S-transferase omega 1 |
6313 |
0.13 |
| chr15_96870341_96870527 | 0.03 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
89 |
0.95 |
| chr12_49763265_49763746 | 0.03 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
2175 |
0.22 |
| chr6_31804565_31804904 | 0.02 |
C6orf48 |
chromosome 6 open reading frame 48 |
114 |
0.47 |
| chr8_145561472_145561771 | 0.02 |
SCRT1 |
scratch family zinc finger 1 |
1678 |
0.16 |
| chr3_111584178_111584329 | 0.02 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
5610 |
0.29 |
| chr20_53950546_53950697 | 0.02 |
ENSG00000252089 |
. |
474270 |
0.01 |
| chr7_116313737_116313903 | 0.02 |
MET |
met proto-oncogene |
1361 |
0.53 |
| chr14_50098662_50099178 | 0.02 |
RP11-649E7.7 |
|
2312 |
0.13 |
| chr5_71310437_71310588 | 0.02 |
MAP1B |
microtubule-associated protein 1B |
92549 |
0.09 |
| chr18_3305605_3305756 | 0.02 |
MYL12B |
myosin, light chain 12B, regulatory |
42726 |
0.12 |
| chr17_60954301_60954474 | 0.02 |
ENSG00000207552 |
. |
67189 |
0.11 |
| chr8_1113646_1113943 | 0.02 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
280466 |
0.01 |
| chr6_27463328_27463580 | 0.02 |
ZNF184 |
zinc finger protein 184 |
22557 |
0.21 |
| chr1_234613311_234613591 | 0.02 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
1398 |
0.49 |
| chr12_54120989_54121200 | 0.02 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
118 |
0.96 |
| chr9_72635619_72635770 | 0.02 |
MAMDC2 |
MAM domain containing 2 |
22803 |
0.23 |
| chr13_38172411_38172686 | 0.02 |
POSTN |
periostin, osteoblast specific factor |
315 |
0.95 |
| chr17_55655751_55655910 | 0.02 |
RP11-118E18.4 |
|
29943 |
0.16 |
| chr12_66221036_66221187 | 0.02 |
HMGA2 |
high mobility group AT-hook 2 |
2208 |
0.34 |
| chr16_87552092_87552315 | 0.02 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
26552 |
0.17 |
| chr13_109639795_109639946 | 0.02 |
MYO16 |
myosin XVI |
101353 |
0.08 |
| chr3_156893095_156894129 | 0.02 |
ENSG00000243176 |
. |
476 |
0.72 |
| chr2_227050440_227050625 | 0.02 |
ENSG00000263363 |
. |
472977 |
0.01 |
| chr12_27399648_27399799 | 0.02 |
STK38L |
serine/threonine kinase 38 like |
1133 |
0.61 |
| chr5_107041117_107041487 | 0.02 |
ENSG00000239708 |
. |
29465 |
0.21 |
| chr6_42192493_42192832 | 0.02 |
MRPS10 |
mitochondrial ribosomal protein S10 |
7059 |
0.19 |
| chr3_194293558_194293709 | 0.02 |
TMEM44-AS1 |
TMEM44 antisense RNA 1 |
11107 |
0.15 |
| chr1_160368401_160368552 | 0.02 |
VANGL2 |
VANGL planar cell polarity protein 2 |
1900 |
0.24 |
| chr15_60212309_60212460 | 0.02 |
FOXB1 |
forkhead box B1 |
84037 |
0.1 |
| chrX_83325612_83325763 | 0.02 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
117228 |
0.07 |
| chr8_8745763_8746197 | 0.02 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
5175 |
0.22 |
| chr1_107682797_107683388 | 0.02 |
NTNG1 |
netrin G1 |
350 |
0.93 |
| chr9_139780499_139780693 | 0.02 |
TRAF2 |
TNF receptor-associated factor 2 |
79 |
0.86 |
| chr7_151123543_151123787 | 0.02 |
RP4-555L14.4 |
|
1104 |
0.36 |
| chr12_122984366_122984751 | 0.02 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
237 |
0.91 |
| chr2_200335677_200335876 | 0.02 |
SATB2 |
SATB homeobox 2 |
213 |
0.91 |
| chr2_190014978_190015129 | 0.02 |
ENSG00000264725 |
. |
17216 |
0.2 |
| chr11_34938171_34938707 | 0.02 |
PDHX |
pyruvate dehydrogenase complex, component X |
251 |
0.64 |
| chr11_72019545_72019702 | 0.02 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
5 |
0.97 |
| chr10_50969933_50970461 | 0.02 |
OGDHL |
oxoglutarate dehydrogenase-like |
172 |
0.97 |
| chr2_66663569_66663965 | 0.02 |
MEIS1 |
Meis homeobox 1 |
758 |
0.57 |
| chrX_73833546_73833857 | 0.02 |
RLIM |
ring finger protein, LIM domain interacting |
751 |
0.74 |
| chr16_2654273_2654460 | 0.02 |
AC141586.5 |
|
891 |
0.35 |
| chr19_45459212_45459543 | 0.02 |
CLPTM1 |
cleft lip and palate associated transmembrane protein 1 |
562 |
0.53 |
| chr8_41649611_41650018 | 0.02 |
ANK1 |
ankyrin 1, erythrocytic |
5326 |
0.2 |
| chr16_86603613_86603777 | 0.02 |
RP11-463O9.5 |
|
2328 |
0.23 |
| chr3_150479775_150480371 | 0.02 |
SIAH2-AS1 |
SIAH2 antisense RNA 1 |
349 |
0.79 |
| chr3_196282059_196282210 | 0.02 |
WDR53 |
WD repeat domain 53 |
12047 |
0.13 |
| chr7_116141667_116142184 | 0.02 |
CAV2 |
caveolin 2 |
2091 |
0.24 |
| chr1_78471558_78472034 | 0.02 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
1258 |
0.34 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |