Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
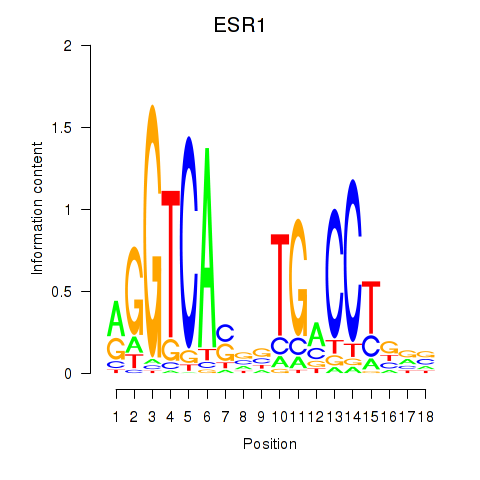
Results for ESR1
Z-value: 0.83
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.17 | estrogen receptor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_152238268_152238543 | ESR1 | 36613 | 0.213069 | 0.67 | 4.8e-02 | Click! |
| chr6_152237063_152237214 | ESR1 | 35346 | 0.216886 | 0.63 | 6.9e-02 | Click! |
| chr6_152043262_152043413 | ESR1 | 31706 | 0.208973 | -0.50 | 1.8e-01 | Click! |
| chr6_151990290_151990441 | ESR1 | 21266 | 0.222650 | 0.47 | 2.1e-01 | Click! |
| chr6_152126783_152127038 | ESR1 | 102 | 0.976307 | 0.45 | 2.2e-01 | Click! |
Activity of the ESR1 motif across conditions
Conditions sorted by the z-value of the ESR1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_11323568_11323719 | 0.36 |
FAM167A |
family with sequence similarity 167, member A |
633 |
0.67 |
| chr13_114831006_114831265 | 0.32 |
RASA3 |
RAS p21 protein activator 3 |
12303 |
0.24 |
| chr20_18037263_18037961 | 0.28 |
OVOL2 |
ovo-like zinc finger 2 |
909 |
0.48 |
| chr3_49940788_49940939 | 0.27 |
MST1R |
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
179 |
0.78 |
| chr11_67078749_67078900 | 0.25 |
SSH3 |
slingshot protein phosphatase 3 |
3696 |
0.13 |
| chr1_153587674_153587825 | 0.25 |
S100A14 |
S100 calcium binding protein A14 |
665 |
0.44 |
| chr22_46482810_46483184 | 0.25 |
FLJ27365 |
hsa-mir-4763 |
1114 |
0.31 |
| chr7_45002030_45002849 | 0.25 |
RP4-647J21.1 |
|
1931 |
0.25 |
| chr14_105435453_105435767 | 0.25 |
AHNAK2 |
AHNAK nucleoprotein 2 |
9084 |
0.16 |
| chr9_110249313_110249729 | 0.24 |
KLF4 |
Kruppel-like factor 4 (gut) |
1890 |
0.38 |
| chr8_10086913_10087064 | 0.24 |
MSRA |
methionine sulfoxide reductase A |
72057 |
0.11 |
| chr8_142289533_142290316 | 0.24 |
RP11-10J21.3 |
Uncharacterized protein |
25260 |
0.12 |
| chr20_62168477_62168628 | 0.24 |
PTK6 |
protein tyrosine kinase 6 |
143 |
0.91 |
| chr9_140708937_140709088 | 0.23 |
ENSG00000207693 |
. |
23859 |
0.19 |
| chr19_39227082_39227399 | 0.23 |
CAPN12 |
calpain 12 |
1178 |
0.28 |
| chr5_1931344_1931629 | 0.23 |
CTD-2194D22.4 |
|
44040 |
0.13 |
| chr14_24630498_24631577 | 0.22 |
IRF9 |
interferon regulatory factor 9 |
527 |
0.47 |
| chr14_91882732_91883366 | 0.22 |
CCDC88C |
coiled-coil domain containing 88C |
641 |
0.78 |
| chr19_41882652_41883217 | 0.22 |
TMEM91 |
transmembrane protein 91 |
60 |
0.74 |
| chr10_13143957_13144489 | 0.22 |
OPTN |
optineurin |
1998 |
0.3 |
| chr5_1315874_1316496 | 0.22 |
ENSG00000263670 |
. |
6693 |
0.18 |
| chr22_20008729_20009650 | 0.22 |
TANGO2 |
transport and golgi organization 2 homolog (Drosophila) |
511 |
0.64 |
| chr18_47089345_47089666 | 0.22 |
LIPG |
lipase, endothelial |
790 |
0.6 |
| chr1_36809017_36809168 | 0.22 |
STK40 |
serine/threonine kinase 40 |
17849 |
0.12 |
| chr1_6531968_6532119 | 0.22 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
5788 |
0.12 |
| chr19_51455517_51455668 | 0.21 |
KLK5 |
kallikrein-related peptidase 5 |
606 |
0.48 |
| chr15_101774475_101774626 | 0.21 |
CHSY1 |
chondroitin sulfate synthase 1 |
17587 |
0.17 |
| chr19_8554749_8554900 | 0.21 |
HNRNPM |
heterogeneous nuclear ribonucleoprotein M |
3782 |
0.14 |
| chr17_48601050_48601201 | 0.21 |
MYCBPAP |
MYCBP associated protein |
44 |
0.95 |
| chr2_74668653_74669044 | 0.21 |
RTKN |
rhotekin |
178 |
0.86 |
| chr17_74868821_74868990 | 0.21 |
MGAT5B |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
176 |
0.95 |
| chr14_100046170_100046321 | 0.21 |
CCDC85C |
coiled-coil domain containing 85C |
218 |
0.93 |
| chr19_2275585_2275736 | 0.21 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
5370 |
0.09 |
| chr4_715290_715441 | 0.20 |
RP11-1191J2.4 |
|
2458 |
0.16 |
| chr4_1796827_1797107 | 0.20 |
FGFR3 |
fibroblast growth factor receptor 3 |
1344 |
0.42 |
| chr20_61733088_61733940 | 0.20 |
ENSG00000225978 |
. |
31 |
0.98 |
| chr20_39970036_39970187 | 0.20 |
LPIN3 |
lipin 3 |
551 |
0.77 |
| chr17_7942417_7942568 | 0.20 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
18 |
0.95 |
| chr2_183225036_183225187 | 0.20 |
ENSG00000242121 |
. |
46100 |
0.17 |
| chr1_3620270_3620421 | 0.20 |
TP73 |
tumor protein p73 |
5723 |
0.12 |
| chr1_26680354_26680505 | 0.20 |
AIM1L |
absent in melanoma 1-like |
192 |
0.9 |
| chr21_41095530_41095681 | 0.20 |
IGSF5 |
immunoglobulin superfamily, member 5 |
21729 |
0.21 |
| chr20_43970794_43971114 | 0.20 |
SDC4 |
syndecan 4 |
6110 |
0.13 |
| chr22_44568627_44569086 | 0.20 |
PARVG |
parvin, gamma |
20 |
0.99 |
| chr1_1015485_1015798 | 0.20 |
RNF223 |
ring finger protein 223 |
5954 |
0.1 |
| chr8_145012042_145012193 | 0.20 |
PLEC |
plectin |
1641 |
0.24 |
| chr8_144820927_144821078 | 0.20 |
FAM83H |
family with sequence similarity 83, member H |
5031 |
0.09 |
| chr22_46546744_46547896 | 0.19 |
PPARA |
peroxisome proliferator-activated receptor alpha |
467 |
0.75 |
| chr14_54430258_54430823 | 0.19 |
BMP4 |
bone morphogenetic protein 4 |
5061 |
0.28 |
| chr20_55204347_55205219 | 0.19 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
425 |
0.81 |
| chr19_54057819_54058678 | 0.19 |
ZNF331 |
zinc finger protein 331 |
167 |
0.9 |
| chr3_5055440_5055591 | 0.19 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
33869 |
0.15 |
| chr1_156092284_156092708 | 0.19 |
LMNA |
lamin A/C |
3455 |
0.14 |
| chr19_10406663_10406906 | 0.19 |
ICAM5 |
intercellular adhesion molecule 5, telencephalin |
5131 |
0.08 |
| chr16_70740729_70741027 | 0.19 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
6730 |
0.15 |
| chr21_33247526_33247677 | 0.19 |
HUNK |
hormonally up-regulated Neu-associated kinase |
1973 |
0.45 |
| chr19_35608806_35608957 | 0.18 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
50 |
0.95 |
| chr12_67807670_67807821 | 0.18 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
114993 |
0.07 |
| chr17_76967334_76967485 | 0.18 |
LGALS3BP |
lectin, galactoside-binding, soluble, 3 binding protein |
5996 |
0.16 |
| chr18_74331565_74332218 | 0.18 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
91017 |
0.08 |
| chr11_66829839_66829990 | 0.18 |
RHOD |
ras homolog family member D |
5554 |
0.17 |
| chr17_46127912_46128198 | 0.18 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
1561 |
0.2 |
| chr1_1173779_1173930 | 0.18 |
B3GALT6 |
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
6225 |
0.08 |
| chr6_96455538_96455689 | 0.18 |
FUT9 |
fucosyltransferase 9 (alpha (1,3) fucosyltransferase) |
8247 |
0.33 |
| chr11_70963000_70963203 | 0.18 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
522 |
0.86 |
| chr16_31146277_31146629 | 0.18 |
PRSS8 |
protease, serine, 8 |
508 |
0.52 |
| chr14_35871775_35873043 | 0.18 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
937 |
0.68 |
| chr11_1552591_1552742 | 0.18 |
DUSP8 |
dual specificity phosphatase 8 |
34500 |
0.1 |
| chr13_106939337_106939488 | 0.18 |
ENSG00000222682 |
. |
131573 |
0.05 |
| chr9_139776272_139777152 | 0.18 |
TRAF2 |
TNF receptor-associated factor 2 |
327 |
0.72 |
| chr14_99710276_99710624 | 0.18 |
AL109767.1 |
|
18835 |
0.2 |
| chr3_10858354_10858505 | 0.18 |
SLC6A11 |
solute carrier family 6 (neurotransmitter transporter), member 11 |
543 |
0.85 |
| chr1_37943283_37943786 | 0.18 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
3381 |
0.18 |
| chr2_232537261_232537412 | 0.17 |
ENSG00000239202 |
. |
26352 |
0.14 |
| chr17_70114398_70114549 | 0.17 |
SOX9 |
SRY (sex determining region Y)-box 9 |
2688 |
0.41 |
| chr2_239036083_239036322 | 0.17 |
ESPNL |
espin-like |
166 |
0.94 |
| chr7_55253774_55253939 | 0.17 |
EGFR-AS1 |
EGFR antisense RNA 1 |
2771 |
0.38 |
| chr2_241949003_241950045 | 0.17 |
AC093585.6 |
|
637 |
0.62 |
| chr16_75416401_75416552 | 0.17 |
RP11-252K23.2 |
|
1318 |
0.36 |
| chr19_1469984_1470717 | 0.17 |
ENSG00000267317 |
. |
7586 |
0.07 |
| chr9_134460250_134460544 | 0.17 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
36828 |
0.13 |
| chr2_239037206_239037357 | 0.17 |
ESPNL |
espin-like |
1245 |
0.38 |
| chr14_55035520_55036036 | 0.17 |
SAMD4A |
sterile alpha motif domain containing 4A |
1141 |
0.56 |
| chrX_48323916_48324124 | 0.17 |
SLC38A5 |
solute carrier family 38, member 5 |
1860 |
0.23 |
| chr1_21620629_21620823 | 0.17 |
ECE1 |
endothelin converting enzyme 1 |
191 |
0.89 |
| chr12_2403889_2404040 | 0.17 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
25022 |
0.23 |
| chr3_52491643_52491832 | 0.17 |
NISCH |
nischarin |
2104 |
0.18 |
| chr7_1690037_1690188 | 0.17 |
ELFN1 |
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
37643 |
0.14 |
| chr6_41703786_41703937 | 0.17 |
TFEB |
transcription factor EB |
136 |
0.93 |
| chr8_144907347_144907498 | 0.17 |
PUF60 |
poly-U binding splicing factor 60KDa |
3774 |
0.11 |
| chr19_34216636_34216787 | 0.17 |
CHST8 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8 |
41277 |
0.18 |
| chr10_125852855_125853389 | 0.17 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
84 |
0.98 |
| chr1_151579139_151579290 | 0.17 |
SNX27 |
sorting nexin family member 27 |
5344 |
0.12 |
| chr6_36098764_36099262 | 0.17 |
MAPK13 |
mitogen-activated protein kinase 13 |
573 |
0.74 |
| chr1_1978273_1978424 | 0.17 |
RP11-547D24.3 |
|
2742 |
0.18 |
| chr1_46955506_46955688 | 0.17 |
DMBX1 |
diencephalon/mesencephalon homeobox 1 |
17071 |
0.16 |
| chr4_1798938_1799089 | 0.17 |
FGFR3 |
fibroblast growth factor receptor 3 |
3390 |
0.22 |
| chr20_1309605_1309756 | 0.17 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
203 |
0.91 |
| chr20_39969728_39969901 | 0.17 |
LPIN3 |
lipin 3 |
254 |
0.92 |
| chr18_74798828_74799607 | 0.16 |
MBP |
myelin basic protein |
18000 |
0.27 |
| chr8_144302737_144302888 | 0.16 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
7744 |
0.14 |
| chr13_24270271_24270700 | 0.16 |
AL139080.1 |
Uncharacterized protein |
51758 |
0.14 |
| chr5_2749613_2749799 | 0.16 |
IRX2 |
iroquois homeobox 2 |
2063 |
0.35 |
| chr11_61657871_61658132 | 0.16 |
FADS3 |
fatty acid desaturase 3 |
852 |
0.49 |
| chr1_55518873_55519024 | 0.16 |
PCSK9 |
proprotein convertase subtilisin/kexin type 9 |
13053 |
0.2 |
| chr19_54369142_54369556 | 0.16 |
MYADM |
myeloid-associated differentiation marker |
128 |
0.89 |
| chr19_13957589_13958095 | 0.16 |
ENSG00000207980 |
. |
10369 |
0.08 |
| chr18_77938309_77938600 | 0.16 |
AC139100.3 |
|
3109 |
0.25 |
| chr16_85832192_85833030 | 0.16 |
COX4I1 |
cytochrome c oxidase subunit IV isoform 1 |
372 |
0.56 |
| chr8_10590631_10590782 | 0.16 |
SOX7 |
SRY (sex determining region Y)-box 7 |
2684 |
0.21 |
| chr1_16538998_16539149 | 0.16 |
ARHGEF19 |
Rho guanine nucleotide exchange factor (GEF) 19 |
31 |
0.96 |
| chr10_35060387_35060538 | 0.16 |
PARD3 |
par-3 family cell polarity regulator |
43787 |
0.16 |
| chr8_145008502_145008738 | 0.16 |
PLEC |
plectin |
5138 |
0.12 |
| chr11_10695962_10696877 | 0.16 |
MRVI1 |
murine retrovirus integration site 1 homolog |
18642 |
0.17 |
| chr10_73769586_73770005 | 0.16 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
45672 |
0.15 |
| chr1_185528978_185529129 | 0.16 |
ENSG00000207108 |
. |
70607 |
0.11 |
| chr17_79378256_79378407 | 0.16 |
ENSG00000266392 |
. |
3753 |
0.14 |
| chr19_11252844_11253247 | 0.16 |
SPC24 |
SPC24, NDC80 kinetochore complex component |
13392 |
0.13 |
| chr3_133476385_133476536 | 0.16 |
TF |
transferrin |
11223 |
0.18 |
| chr4_3043146_3044167 | 0.16 |
ENSG00000199335 |
. |
2172 |
0.29 |
| chr7_139047352_139047690 | 0.16 |
C7orf55-LUC7L2 |
C7orf55-LUC7L2 readthrough |
2867 |
0.19 |
| chr20_45946936_45948261 | 0.16 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
352 |
0.82 |
| chr2_149895781_149895992 | 0.16 |
LYPD6B |
LY6/PLAUR domain containing 6B |
654 |
0.82 |
| chr10_29978497_29978648 | 0.15 |
ENSG00000222092 |
. |
22272 |
0.21 |
| chr19_3387681_3387832 | 0.15 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
21172 |
0.16 |
| chr16_57653725_57653876 | 0.15 |
GPR56 |
G protein-coupled receptor 56 |
54 |
0.97 |
| chr4_17783377_17783652 | 0.15 |
FAM184B |
family with sequence similarity 184, member B |
379 |
0.88 |
| chr22_19792099_19792250 | 0.15 |
TBX1 |
T-box 1 |
47948 |
0.1 |
| chr8_22423482_22424350 | 0.15 |
SORBS3 |
sorbin and SH3 domain containing 3 |
402 |
0.6 |
| chr8_145698019_145698220 | 0.15 |
FOXH1 |
forkhead box H1 |
3599 |
0.07 |
| chr5_58055562_58056142 | 0.15 |
RP11-479O16.1 |
|
24754 |
0.24 |
| chr2_106886104_106886958 | 0.15 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
75736 |
0.1 |
| chr19_13961881_13962048 | 0.15 |
ENSG00000207980 |
. |
14491 |
0.07 |
| chr6_50817384_50817535 | 0.15 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
31020 |
0.25 |
| chr8_128919529_128920109 | 0.15 |
TMEM75 |
transmembrane protein 75 |
40772 |
0.15 |
| chr22_40391518_40391669 | 0.15 |
FAM83F |
family with sequence similarity 83, member F |
640 |
0.7 |
| chr19_940940_941628 | 0.15 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
8775 |
0.08 |
| chr6_160721267_160721418 | 0.15 |
SLC22A2 |
solute carrier family 22 (organic cation transporter), member 2 |
22672 |
0.23 |
| chr11_65360517_65360668 | 0.15 |
KCNK7 |
potassium channel, subfamily K, member 7 |
380 |
0.52 |
| chr1_34631906_34632332 | 0.15 |
C1orf94 |
chromosome 1 open reading frame 94 |
365 |
0.8 |
| chr17_15554007_15555167 | 0.15 |
RP11-640I15.1 |
|
317 |
0.52 |
| chr5_55839563_55839714 | 0.15 |
AC022431.3 |
|
5985 |
0.25 |
| chr12_125002282_125002834 | 0.15 |
NCOR2 |
nuclear receptor corepressor 2 |
282 |
0.95 |
| chr17_79779492_79779643 | 0.15 |
FAM195B |
family with sequence similarity 195, member B |
2796 |
0.11 |
| chr19_4061941_4062216 | 0.15 |
CTD-2622I13.3 |
|
669 |
0.55 |
| chr11_60609533_60610326 | 0.15 |
CCDC86 |
coiled-coil domain containing 86 |
385 |
0.51 |
| chr16_3014465_3014677 | 0.15 |
KREMEN2 |
kringle containing transmembrane protein 2 |
49 |
0.93 |
| chr1_41966637_41966788 | 0.15 |
EDN2 |
endothelin 2 |
16370 |
0.19 |
| chr1_12675825_12676469 | 0.15 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
1590 |
0.37 |
| chr17_8218792_8218943 | 0.15 |
AC135178.7 |
|
3163 |
0.11 |
| chr19_49567989_49568490 | 0.15 |
NTF4 |
neurotrophin 4 |
72 |
0.9 |
| chr17_61513067_61513218 | 0.15 |
CYB561 |
cytochrome b561 |
597 |
0.65 |
| chr9_103791551_103791702 | 0.15 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
635 |
0.84 |
| chr22_46959420_46959714 | 0.15 |
GRAMD4 |
GRAM domain containing 4 |
12342 |
0.19 |
| chr19_55658159_55658310 | 0.15 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
76 |
0.93 |
| chr14_105330590_105330952 | 0.15 |
CEP170B |
centrosomal protein 170B |
846 |
0.6 |
| chr11_6341290_6341712 | 0.15 |
PRKCDBP |
protein kinase C, delta binding protein |
261 |
0.9 |
| chr20_62573957_62574108 | 0.14 |
ENSG00000207554 |
. |
47 |
0.9 |
| chr17_7492081_7492232 | 0.14 |
SOX15 |
SRY (sex determining region Y)-box 15 |
1234 |
0.16 |
| chr11_8299597_8299748 | 0.14 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
9409 |
0.26 |
| chr20_39945349_39946123 | 0.14 |
ZHX3 |
zinc fingers and homeoboxes 3 |
90 |
0.97 |
| chr5_150970622_150970773 | 0.14 |
FAT2 |
FAT atypical cadherin 2 |
22192 |
0.17 |
| chrX_153187182_153187333 | 0.14 |
ARHGAP4 |
Rho GTPase activating protein 4 |
369 |
0.73 |
| chr17_43249924_43250075 | 0.14 |
RP13-890H12.2 |
|
1048 |
0.34 |
| chr1_155097548_155097699 | 0.14 |
EFNA1 |
ephrin-A1 |
2313 |
0.11 |
| chr5_167243776_167243927 | 0.14 |
ENSG00000221741 |
. |
53819 |
0.13 |
| chr8_145020311_145020609 | 0.14 |
PLEC |
plectin |
113 |
0.91 |
| chr19_3098811_3099227 | 0.14 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
4611 |
0.13 |
| chr14_105564128_105564279 | 0.14 |
GPR132 |
G protein-coupled receptor 132 |
32421 |
0.14 |
| chr19_1749690_1749939 | 0.14 |
ONECUT3 |
one cut homeobox 3 |
2558 |
0.2 |
| chrX_11782921_11783072 | 0.14 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
5249 |
0.33 |
| chr6_12288567_12288718 | 0.14 |
EDN1 |
endothelin 1 |
1954 |
0.48 |
| chr17_79316804_79317024 | 0.14 |
TMEM105 |
transmembrane protein 105 |
12440 |
0.13 |
| chr1_156785806_156786457 | 0.14 |
SH2D2A |
SH2 domain containing 2A |
495 |
0.51 |
| chr13_114186665_114186816 | 0.14 |
TMCO3 |
transmembrane and coiled-coil domains 3 |
41382 |
0.11 |
| chr1_153507238_153508458 | 0.14 |
S100A6 |
S100 calcium binding protein A6 |
619 |
0.48 |
| chr1_6479504_6479655 | 0.14 |
HES2 |
hes family bHLH transcription factor 2 |
384 |
0.76 |
| chr11_1410043_1410900 | 0.14 |
BRSK2 |
BR serine/threonine kinase 2 |
658 |
0.72 |
| chr16_89427901_89428186 | 0.14 |
ANKRD11 |
ankyrin repeat domain 11 |
32649 |
0.1 |
| chr16_68824059_68824234 | 0.14 |
RP11-354M1.2 |
|
33283 |
0.12 |
| chr4_1761839_1761990 | 0.14 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
31783 |
0.12 |
| chr1_230223344_230223516 | 0.14 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20412 |
0.23 |
| chr2_120041666_120041961 | 0.14 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
35166 |
0.15 |
| chr11_567710_568184 | 0.14 |
ENSG00000199038 |
. |
251 |
0.8 |
| chr19_42440673_42440856 | 0.14 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
13518 |
0.12 |
| chr10_99331899_99332050 | 0.14 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
224 |
0.89 |
| chr8_142083960_142084111 | 0.14 |
DENND3 |
DENN/MADD domain containing 3 |
43342 |
0.14 |
| chr17_78866297_78866787 | 0.14 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
29982 |
0.14 |
| chr19_13110560_13110829 | 0.14 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
4042 |
0.11 |
| chr11_3876946_3878257 | 0.14 |
STIM1 |
stromal interaction molecule 1 |
189 |
0.59 |
| chr15_31757911_31758062 | 0.14 |
KLF13 |
Kruppel-like factor 13 |
99629 |
0.08 |
| chr17_7256192_7256343 | 0.14 |
KCTD11 |
potassium channel tetramerization domain containing 11 |
1059 |
0.2 |
| chr6_108141455_108142294 | 0.14 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
3642 |
0.33 |
| chr7_73021530_73021732 | 0.14 |
MLXIPL |
MLX interacting protein-like |
8860 |
0.14 |
| chr2_128395783_128395934 | 0.14 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
3848 |
0.17 |
| chr17_38656484_38656635 | 0.14 |
TNS4 |
tensin 4 |
1290 |
0.42 |
| chr1_38513049_38513583 | 0.14 |
POU3F1 |
POU class 3 homeobox 1 |
866 |
0.54 |
| chr9_131126990_131127303 | 0.14 |
URM1 |
ubiquitin related modifier 1 |
6452 |
0.1 |
| chr10_8076986_8077137 | 0.14 |
GATA3 |
GATA binding protein 3 |
19595 |
0.29 |
| chr13_30168298_30169638 | 0.14 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
857 |
0.74 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 0.3 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.1 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.2 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.1 | 0.2 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.2 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.1 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.3 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.1 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.1 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.1 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.2 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.2 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.4 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.2 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.2 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0061339 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.2 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0001839 | neural plate morphogenesis(GO:0001839) neural plate development(GO:0001840) |
| 0.0 | 0.1 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0010984 | regulation of lipoprotein particle clearance(GO:0010984) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.0 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0032610 | interleukin-1 alpha production(GO:0032610) |
| 0.0 | 0.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0055026 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.0 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:1901862 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.0 | 0.0 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.2 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0030850 | prostate gland development(GO:0030850) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.1 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.0 | GO:0042634 | regulation of hair cycle(GO:0042634) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.0 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.0 | GO:0002363 | alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.0 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.0 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.0 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.0 | 0.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.0 | GO:0002830 | positive regulation of type 2 immune response(GO:0002830) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.0 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.2 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |