Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
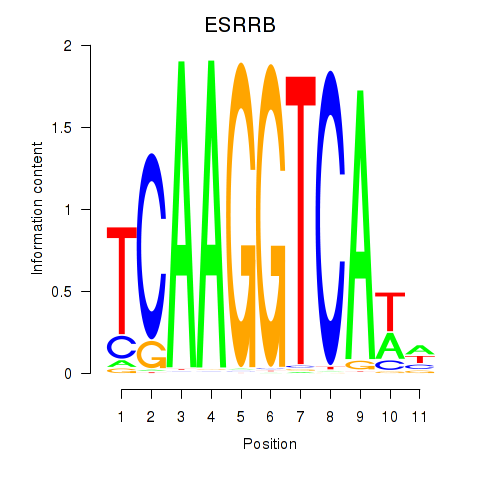
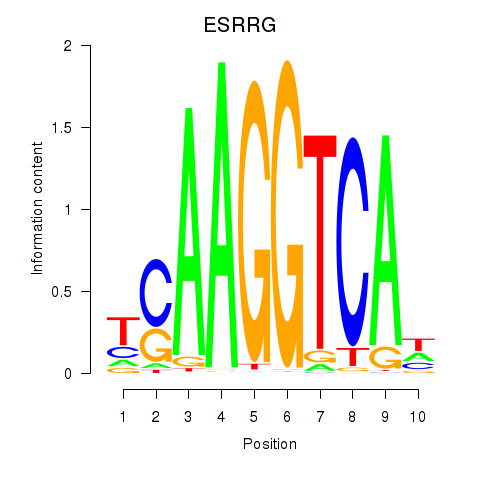
Results for ESRRB_ESRRG
Z-value: 0.42
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.10 | estrogen related receptor beta |
|
ESRRG
|
ENSG00000196482.12 | estrogen related receptor gamma |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_76789957_76790108 | ESRRB | 13075 | 0.190064 | -0.78 | 1.3e-02 | Click! |
| chr14_77012691_77012842 | ESRRB | 138885 | 0.044035 | -0.59 | 9.2e-02 | Click! |
| chr14_77017371_77017522 | ESRRB | 143565 | 0.041560 | -0.45 | 2.3e-01 | Click! |
| chr14_76939966_76940117 | ESRRB | 66160 | 0.122724 | 0.14 | 7.1e-01 | Click! |
| chr14_77013139_77013290 | ESRRB | 139333 | 0.043794 | -0.13 | 7.4e-01 | Click! |
| chr1_217326385_217326536 | ESRRG | 15363 | 0.311735 | -0.61 | 7.9e-02 | Click! |
| chr1_217076565_217076716 | ESRRG | 36372 | 0.243772 | -0.56 | 1.2e-01 | Click! |
| chr1_216796230_216796381 | ESRRG | 100369 | 0.086990 | -0.53 | 1.4e-01 | Click! |
| chr1_217113777_217113928 | ESRRG | 837 | 0.779096 | -0.43 | 2.5e-01 | Click! |
| chr1_217448442_217448593 | ESRRG | 137420 | 0.052700 | -0.36 | 3.4e-01 | Click! |
Activity of the ESRRB_ESRRG motif across conditions
Conditions sorted by the z-value of the ESRRB_ESRRG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
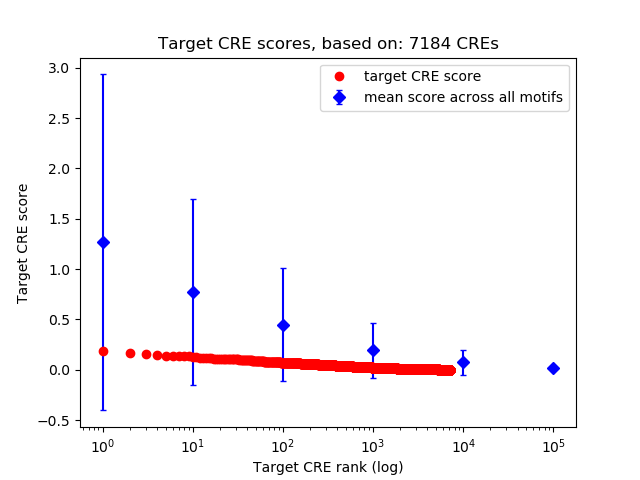
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_24024495_24024646 | 0.19 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
16729 |
0.28 |
| chr9_139607462_139607817 | 0.17 |
FAM69B |
family with sequence similarity 69, member B |
617 |
0.49 |
| chr1_71230340_71230491 | 0.15 |
RP3-333A15.1 |
|
182647 |
0.03 |
| chr16_22307896_22308092 | 0.14 |
POLR3E |
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
736 |
0.63 |
| chr5_88178054_88178448 | 0.14 |
MEF2C |
myocyte enhancer factor 2C |
713 |
0.52 |
| chr11_85521285_85521488 | 0.14 |
SYTL2 |
synaptotagmin-like 2 |
786 |
0.67 |
| chr5_175085682_175085863 | 0.14 |
HRH2 |
histamine receptor H2 |
739 |
0.7 |
| chr20_4202584_4202789 | 0.13 |
ADRA1D |
adrenoceptor alpha 1D |
27035 |
0.17 |
| chr21_40051384_40051625 | 0.13 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
17800 |
0.28 |
| chr3_187989024_187989175 | 0.13 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
31447 |
0.22 |
| chr18_13459821_13459972 | 0.13 |
ENSG00000266146 |
. |
50 |
0.95 |
| chr6_143508899_143509050 | 0.12 |
AIG1 |
androgen-induced 1 |
61587 |
0.13 |
| chr16_70718792_70718985 | 0.12 |
MTSS1L |
metastasis suppressor 1-like |
1081 |
0.43 |
| chr19_39108076_39108634 | 0.12 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
209 |
0.87 |
| chr5_32734660_32734811 | 0.12 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
22298 |
0.21 |
| chr6_157297379_157297598 | 0.11 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
74981 |
0.12 |
| chr18_44497626_44498111 | 0.11 |
PIAS2 |
protein inhibitor of activated STAT, 2 |
136 |
0.79 |
| chr10_121045220_121045419 | 0.11 |
RP11-79M19.2 |
|
50375 |
0.11 |
| chr9_21481557_21481708 | 0.11 |
IFNE |
interferon, epsilon |
680 |
0.64 |
| chr2_191915311_191915462 | 0.11 |
ENSG00000231858 |
. |
29134 |
0.15 |
| chr1_23493687_23494408 | 0.11 |
LUZP1 |
leucine zipper protein 1 |
1263 |
0.35 |
| chr15_89676296_89676447 | 0.11 |
ENSG00000239151 |
. |
12361 |
0.18 |
| chr8_105450624_105450775 | 0.11 |
ENSG00000266734 |
. |
2925 |
0.23 |
| chr5_175962769_175963088 | 0.11 |
RNF44 |
ring finger protein 44 |
1493 |
0.28 |
| chr7_38315182_38315333 | 0.11 |
STARD3NL |
STARD3 N-terminal like |
97260 |
0.09 |
| chr11_86666736_86666936 | 0.11 |
FZD4 |
frizzled family receptor 4 |
403 |
0.89 |
| chr18_479598_479768 | 0.10 |
COLEC12 |
collectin sub-family member 12 |
21039 |
0.18 |
| chr1_160680341_160680580 | 0.10 |
CD48 |
CD48 molecule |
1133 |
0.45 |
| chr13_40359180_40359331 | 0.10 |
ENSG00000200526 |
. |
392 |
0.9 |
| chr9_138998645_138998796 | 0.10 |
C9orf69 |
chromosome 9 open reading frame 69 |
11400 |
0.19 |
| chr9_80524074_80524374 | 0.10 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chr2_237445797_237446029 | 0.10 |
IQCA1 |
IQ motif containing with AAA domain 1 |
29728 |
0.19 |
| chr16_70559995_70560146 | 0.10 |
SF3B3 |
splicing factor 3b, subunit 3, 130kDa |
734 |
0.51 |
| chr2_39731663_39731814 | 0.10 |
AC007246.3 |
|
9549 |
0.25 |
| chr11_75041710_75041861 | 0.10 |
ENSG00000199090 |
. |
4445 |
0.17 |
| chr3_161499334_161499485 | 0.10 |
OTOL1 |
otolin 1 |
284813 |
0.01 |
| chr8_42434388_42434539 | 0.10 |
SMIM19 |
small integral membrane protein 19 |
32892 |
0.14 |
| chr19_11590516_11590718 | 0.10 |
ELAVL3 |
ELAV like neuron-specific RNA binding protein 3 |
823 |
0.4 |
| chr20_47784288_47784439 | 0.10 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
6437 |
0.22 |
| chr22_39843892_39844043 | 0.10 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
9382 |
0.15 |
| chr12_118796178_118796667 | 0.09 |
TAOK3 |
TAO kinase 3 |
488 |
0.85 |
| chr15_57512568_57512719 | 0.09 |
TCF12 |
transcription factor 12 |
979 |
0.67 |
| chr5_1594076_1595069 | 0.09 |
SDHAP3 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3 |
1168 |
0.53 |
| chr3_66101350_66101501 | 0.09 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
17860 |
0.25 |
| chr22_39417668_39418190 | 0.09 |
APOBEC3D |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
472 |
0.69 |
| chr19_16225619_16225770 | 0.09 |
RAB8A |
RAB8A, member RAS oncogene family |
2987 |
0.18 |
| chr17_900716_900867 | 0.09 |
TIMM22 |
translocase of inner mitochondrial membrane 22 homolog (yeast) |
434 |
0.77 |
| chr1_19832105_19832279 | 0.09 |
ENSG00000201609 |
. |
4895 |
0.17 |
| chr7_26410239_26410390 | 0.09 |
AC004540.4 |
|
5578 |
0.24 |
| chr3_113251981_113252391 | 0.09 |
SIDT1 |
SID1 transmembrane family, member 1 |
968 |
0.54 |
| chr2_102431316_102431467 | 0.09 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
17665 |
0.26 |
| chr1_65558220_65558387 | 0.09 |
ENSG00000199135 |
. |
34112 |
0.15 |
| chr16_16046187_16046378 | 0.09 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
2848 |
0.29 |
| chr19_58133868_58134073 | 0.09 |
ZNF211 |
zinc finger protein 211 |
7791 |
0.09 |
| chr2_86080820_86081072 | 0.09 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
13831 |
0.17 |
| chr19_2720102_2720571 | 0.09 |
DIRAS1 |
DIRAS family, GTP-binding RAS-like 1 |
1003 |
0.38 |
| chr20_62477557_62477708 | 0.09 |
AL158091.1 |
Protein LOC100509861 |
2359 |
0.14 |
| chr16_86700893_86701044 | 0.08 |
FOXL1 |
forkhead box L1 |
88853 |
0.08 |
| chr14_96003897_96004048 | 0.08 |
GLRX5 |
glutaredoxin 5 |
2361 |
0.21 |
| chr3_56955767_56956017 | 0.08 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
5393 |
0.25 |
| chr2_198365048_198365752 | 0.08 |
HSPE1-MOB4 |
HSPE1-MOB4 readthrough |
263 |
0.41 |
| chr5_166899834_166899985 | 0.08 |
CTB-105L4.2 |
|
163034 |
0.03 |
| chrX_9310842_9311361 | 0.08 |
TBL1X |
transducin (beta)-like 1X-linked |
120234 |
0.06 |
| chr22_28063586_28063737 | 0.08 |
RP11-375H17.1 |
|
48807 |
0.18 |
| chr3_15838336_15838568 | 0.08 |
ANKRD28 |
ankyrin repeat domain 28 |
424 |
0.86 |
| chr4_89743291_89743450 | 0.08 |
FAM13A |
family with sequence similarity 13, member A |
982 |
0.66 |
| chr3_141516104_141516959 | 0.08 |
ENSG00000265391 |
. |
9778 |
0.19 |
| chr17_76755792_76756034 | 0.08 |
CYTH1 |
cytohesin 1 |
22441 |
0.17 |
| chr21_36401278_36401429 | 0.08 |
RUNX1 |
runt-related transcription factor 1 |
20109 |
0.29 |
| chr5_15621567_15621718 | 0.08 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
5551 |
0.27 |
| chr15_26105615_26105919 | 0.08 |
ATP10A |
ATPase, class V, type 10A |
2588 |
0.28 |
| chr4_87994321_87994472 | 0.08 |
AFF1 |
AF4/FMR2 family, member 1 |
23592 |
0.24 |
| chr5_137843995_137844146 | 0.08 |
ETF1 |
eukaryotic translation termination factor 1 |
33330 |
0.11 |
| chr16_30787280_30787650 | 0.08 |
RP11-2C24.6 |
|
2305 |
0.15 |
| chr12_7064003_7064154 | 0.08 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
3540 |
0.07 |
| chr15_64754210_64754590 | 0.08 |
ZNF609 |
zinc finger protein 609 |
1459 |
0.3 |
| chr16_66967480_66967691 | 0.08 |
FAM96B |
family with sequence similarity 96, member B |
568 |
0.45 |
| chr6_133089264_133089460 | 0.08 |
VNN2 |
vanin 2 |
4764 |
0.14 |
| chr2_122044989_122045140 | 0.08 |
TFCP2L1 |
transcription factor CP2-like 1 |
2281 |
0.44 |
| chr5_43483064_43483215 | 0.08 |
C5orf28 |
chromosome 5 open reading frame 28 |
799 |
0.42 |
| chr15_59730475_59730927 | 0.08 |
FAM81A |
family with sequence similarity 81, member A |
6 |
0.98 |
| chr12_72234485_72234704 | 0.08 |
TBC1D15 |
TBC1 domain family, member 15 |
1036 |
0.63 |
| chrX_117632028_117632179 | 0.07 |
DOCK11 |
dedicator of cytokinesis 11 |
2231 |
0.39 |
| chr10_35658048_35658233 | 0.07 |
CCNY |
cyclin Y |
32338 |
0.17 |
| chr17_56349285_56349436 | 0.07 |
MPO |
myeloperoxidase |
1493 |
0.3 |
| chrX_62781284_62781641 | 0.07 |
ENSG00000235437 |
. |
511 |
0.87 |
| chr19_13044658_13044845 | 0.07 |
FARSA |
phenylalanyl-tRNA synthetase, alpha subunit |
100 |
0.89 |
| chr1_183843426_183843577 | 0.07 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
69211 |
0.11 |
| chr17_19967741_19968028 | 0.07 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
22450 |
0.18 |
| chr10_116156727_116156878 | 0.07 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
7437 |
0.24 |
| chr6_124160195_124160346 | 0.07 |
RP11-374A22.1 |
|
15885 |
0.24 |
| chr2_66672542_66672972 | 0.07 |
MEIS1 |
Meis homeobox 1 |
2660 |
0.22 |
| chr7_157112598_157112749 | 0.07 |
ENSG00000266453 |
. |
14186 |
0.22 |
| chr3_111266223_111266431 | 0.07 |
CD96 |
CD96 molecule |
5330 |
0.28 |
| chr20_20714689_20714840 | 0.07 |
ENSG00000264361 |
. |
4610 |
0.29 |
| chr16_17107310_17107606 | 0.07 |
CTD-2576D5.4 |
|
120903 |
0.07 |
| chr17_55889172_55889323 | 0.07 |
ENSG00000222976 |
. |
22437 |
0.16 |
| chr11_61601877_61602071 | 0.07 |
FADS1 |
fatty acid desaturase 1 |
5184 |
0.12 |
| chr3_44551397_44551548 | 0.07 |
ZNF852 |
zinc finger protein 852 |
656 |
0.63 |
| chr19_29703517_29704012 | 0.07 |
UQCRFS1 |
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
684 |
0.77 |
| chr11_64511299_64511931 | 0.07 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
2 |
0.96 |
| chr20_31169585_31169736 | 0.07 |
RP11-410N8.1 |
|
105 |
0.95 |
| chr1_167904186_167904337 | 0.07 |
MPC2 |
mitochondrial pyruvate carrier 2 |
1017 |
0.45 |
| chr10_63213041_63213324 | 0.07 |
TMEM26 |
transmembrane protein 26 |
26 |
0.95 |
| chr12_50064620_50064771 | 0.07 |
FMNL3 |
formin-like 3 |
36308 |
0.11 |
| chrX_3617821_3618286 | 0.07 |
PRKX |
protein kinase, X-linked |
13596 |
0.21 |
| chr17_41853971_41854122 | 0.07 |
DUSP3 |
dual specificity phosphatase 3 |
1740 |
0.22 |
| chr10_65025121_65025369 | 0.07 |
JMJD1C |
jumonji domain containing 1C |
3581 |
0.34 |
| chr6_97346064_97346321 | 0.07 |
NDUFAF4 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
435 |
0.83 |
| chr22_32341496_32341778 | 0.07 |
C22orf24 |
chromosome 22 open reading frame 24 |
301 |
0.81 |
| chr8_87614320_87614471 | 0.07 |
CNGB3 |
cyclic nucleotide gated channel beta 3 |
22961 |
0.2 |
| chr12_93988003_93988251 | 0.07 |
SOCS2 |
suppressor of cytokine signaling 2 |
19293 |
0.17 |
| chr7_2443371_2444332 | 0.07 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
117 |
0.97 |
| chr17_32573755_32573906 | 0.07 |
CCL2 |
chemokine (C-C motif) ligand 2 |
8474 |
0.13 |
| chr17_72708617_72709026 | 0.07 |
CD300LF |
CD300 molecule-like family member f |
259 |
0.86 |
| chr4_53862383_53862534 | 0.07 |
RP11-752D24.2 |
|
50602 |
0.17 |
| chr22_22178579_22178730 | 0.07 |
ENSG00000200985 |
. |
31931 |
0.12 |
| chr11_33910636_33910787 | 0.07 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
3125 |
0.27 |
| chr3_50656380_50656531 | 0.07 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
1634 |
0.31 |
| chr5_33364481_33364632 | 0.07 |
CTD-2203K17.1 |
|
76169 |
0.11 |
| chr8_14486227_14486378 | 0.07 |
SGCZ |
sarcoglycan, zeta |
73867 |
0.11 |
| chr5_139017553_139017879 | 0.07 |
CXXC5 |
CXXC finger protein 5 |
9168 |
0.21 |
| chr19_54880943_54881230 | 0.07 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
972 |
0.39 |
| chr12_889891_890042 | 0.07 |
ENSG00000221439 |
. |
333 |
0.9 |
| chr11_36112380_36112531 | 0.07 |
ENSG00000263389 |
. |
80807 |
0.09 |
| chr13_41837104_41837614 | 0.07 |
MTRF1 |
mitochondrial translational release factor 1 |
261 |
0.92 |
| chr19_37096291_37096844 | 0.07 |
ZNF382 |
zinc finger protein 382 |
330 |
0.52 |
| chr16_84261621_84261868 | 0.07 |
KCNG4 |
potassium voltage-gated channel, subfamily G, member 4 |
11560 |
0.12 |
| chr2_9403075_9403226 | 0.07 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
56256 |
0.15 |
| chr7_30199656_30199839 | 0.07 |
AC007036.5 |
|
2600 |
0.25 |
| chr4_171418341_171418769 | 0.07 |
ENSG00000251961 |
. |
394063 |
0.01 |
| chr2_206767490_206767641 | 0.07 |
ENSG00000201875 |
. |
2211 |
0.38 |
| chr4_77483255_77483500 | 0.07 |
ENSG00000263445 |
. |
11344 |
0.16 |
| chr2_112209446_112209704 | 0.07 |
ENSG00000266139 |
. |
130907 |
0.06 |
| chr6_31804914_31805281 | 0.07 |
ENSG00000201754 |
. |
244 |
0.6 |
| chr3_99928980_99929378 | 0.07 |
ENSG00000238377 |
. |
11257 |
0.16 |
| chrX_54836533_54836684 | 0.07 |
MAGED2 |
melanoma antigen family D, 2 |
1115 |
0.48 |
| chr10_61142631_61142782 | 0.07 |
FAM13C |
family with sequence similarity 13, member C |
19767 |
0.27 |
| chr7_90315781_90315932 | 0.06 |
CDK14 |
cyclin-dependent kinase 14 |
22724 |
0.24 |
| chr10_22422535_22422686 | 0.06 |
EBLN1 |
endogenous Bornavirus-like nucleoprotein 1 |
76340 |
0.1 |
| chr2_9615136_9615532 | 0.06 |
IAH1 |
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) |
150 |
0.95 |
| chr4_3323886_3324037 | 0.06 |
RGS12 |
regulator of G-protein signaling 12 |
6166 |
0.26 |
| chr4_166251653_166251827 | 0.06 |
MSMO1 |
methylsterol monooxygenase 1 |
2661 |
0.29 |
| chr8_6132048_6132199 | 0.06 |
RP11-115C21.2 |
|
131940 |
0.05 |
| chr5_59542908_59543059 | 0.06 |
ENSG00000202017 |
. |
56995 |
0.15 |
| chr16_63849514_63849665 | 0.06 |
RP11-96H17.3 |
|
748695 |
0.0 |
| chr5_66489318_66489469 | 0.06 |
CD180 |
CD180 molecule |
3234 |
0.35 |
| chr12_57038653_57039701 | 0.06 |
ENSG00000207031 |
. |
292 |
0.67 |
| chr3_183005602_183005777 | 0.06 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
22557 |
0.18 |
| chr1_67218434_67218679 | 0.06 |
TCTEX1D1 |
Tctex1 domain containing 1 |
413 |
0.83 |
| chr17_43663178_43663690 | 0.06 |
RP11-798G7.4 |
|
26998 |
0.11 |
| chr3_183165055_183165319 | 0.06 |
LINC00888 |
long intergenic non-protein coding RNA 888 |
4342 |
0.13 |
| chr19_6481304_6482171 | 0.06 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr10_70821404_70821710 | 0.06 |
SRGN |
serglycin |
26317 |
0.16 |
| chr1_55677031_55677324 | 0.06 |
USP24 |
ubiquitin specific peptidase 24 |
3585 |
0.27 |
| chr8_27262752_27262933 | 0.06 |
PTK2B |
protein tyrosine kinase 2 beta |
7873 |
0.23 |
| chr10_116527243_116527917 | 0.06 |
FAM160B1 |
family with sequence similarity 160, member B1 |
53923 |
0.15 |
| chr15_62851937_62852088 | 0.06 |
TLN2 |
talin 2 |
1552 |
0.51 |
| chr19_54288907_54289058 | 0.06 |
ENSG00000199031 |
. |
1947 |
0.07 |
| chr4_16258000_16258171 | 0.06 |
TAPT1-AS1 |
TAPT1 antisense RNA 1 (head to head) |
29417 |
0.18 |
| chr7_115116914_115117103 | 0.06 |
ENSG00000202377 |
. |
104488 |
0.08 |
| chr19_2235339_2235739 | 0.06 |
PLEKHJ1 |
pleckstrin homology domain containing, family J member 1 |
732 |
0.32 |
| chr5_141693565_141693716 | 0.06 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
10107 |
0.23 |
| chr17_41857566_41857858 | 0.06 |
C17orf105 |
chromosome 17 open reading frame 105 |
91 |
0.93 |
| chr18_44336370_44336521 | 0.06 |
ENSG00000270112 |
. |
5 |
0.84 |
| chr9_117798033_117798184 | 0.06 |
TNC |
tenascin C |
29083 |
0.23 |
| chr8_124775567_124775718 | 0.06 |
FAM91A1 |
family with sequence similarity 91, member A1 |
5054 |
0.24 |
| chr19_55574049_55574487 | 0.06 |
RDH13 |
retinol dehydrogenase 13 (all-trans/9-cis) |
276 |
0.84 |
| chr5_149827841_149827992 | 0.06 |
RPS14 |
ribosomal protein S14 |
560 |
0.75 |
| chr11_20410264_20410454 | 0.06 |
PRMT3 |
protein arginine methyltransferase 3 |
1097 |
0.6 |
| chr6_148521629_148521780 | 0.06 |
SASH1 |
SAM and SH3 domain containing 1 |
71736 |
0.11 |
| chr14_90083201_90083388 | 0.06 |
FOXN3 |
forkhead box N3 |
2180 |
0.23 |
| chr10_88611537_88611688 | 0.06 |
ENSG00000200176 |
. |
37714 |
0.13 |
| chr6_139012958_139013212 | 0.06 |
RP11-390P2.4 |
|
600 |
0.46 |
| chr11_65383666_65384440 | 0.06 |
PCNXL3 |
pecanex-like 3 (Drosophila) |
809 |
0.35 |
| chr20_50727464_50727629 | 0.06 |
ZFP64 |
ZFP64 zinc finger protein |
5363 |
0.34 |
| chr1_193073335_193073486 | 0.06 |
GLRX2 |
glutaredoxin 2 |
1151 |
0.39 |
| chr5_139557686_139557862 | 0.06 |
CYSTM1 |
cysteine-rich transmembrane module containing 1 |
3547 |
0.18 |
| chr6_134571322_134571473 | 0.06 |
ENSG00000200058 |
. |
6776 |
0.19 |
| chr16_88767040_88767399 | 0.06 |
RNF166 |
ring finger protein 166 |
208 |
0.83 |
| chr11_88068071_88068222 | 0.06 |
CTSC |
cathepsin C |
2755 |
0.4 |
| chr12_47583133_47583337 | 0.06 |
ENSG00000263838 |
. |
1640 |
0.43 |
| chr2_204606154_204606338 | 0.06 |
ENSG00000211573 |
. |
23275 |
0.19 |
| chr2_129230436_129230587 | 0.06 |
ENSG00000238379 |
. |
27739 |
0.24 |
| chr1_183445403_183445616 | 0.06 |
SMG7 |
SMG7 nonsense mediated mRNA decay factor |
3442 |
0.27 |
| chr12_52306155_52306306 | 0.06 |
ACVRL1 |
activin A receptor type II-like 1 |
75 |
0.97 |
| chr2_152956097_152956248 | 0.06 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
579 |
0.69 |
| chr8_72087084_72087235 | 0.06 |
RP11-326E22.1 |
|
19402 |
0.28 |
| chr3_113666984_113667185 | 0.06 |
ZDHHC23 |
zinc finger, DHHC-type containing 23 |
262 |
0.78 |
| chr8_18868385_18868536 | 0.06 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
2736 |
0.3 |
| chr14_104023687_104023838 | 0.06 |
RP11-894P9.2 |
|
4004 |
0.09 |
| chr19_3062696_3062847 | 0.06 |
AES |
amino-terminal enhancer of split |
110 |
0.94 |
| chr17_46646430_46646725 | 0.06 |
HOXB3 |
homeobox B3 |
4808 |
0.08 |
| chr17_39265148_39265305 | 0.06 |
KRTAP4-9 |
keratin associated protein 4-9 |
3642 |
0.08 |
| chr1_183829081_183829232 | 0.06 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
54866 |
0.14 |
| chrX_68776626_68776777 | 0.06 |
FAM155B |
family with sequence similarity 155, member B |
51617 |
0.13 |
| chr6_146057180_146057409 | 0.06 |
EPM2A |
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
134 |
0.98 |
| chr5_142382910_142383061 | 0.06 |
ARHGAP26 |
Rho GTPase activating protein 26 |
33776 |
0.23 |
| chr14_73737042_73737350 | 0.06 |
RP4-647C14.3 |
|
3593 |
0.17 |
| chr11_125926321_125926472 | 0.06 |
CDON |
cell adhesion associated, oncogene regulated |
6310 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |