Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
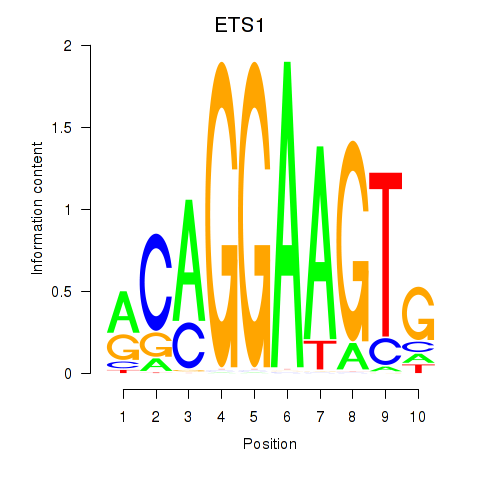
Results for ETS1
Z-value: 1.53
Transcription factors associated with ETS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETS1
|
ENSG00000134954.10 | ETS proto-oncogene 1, transcription factor |
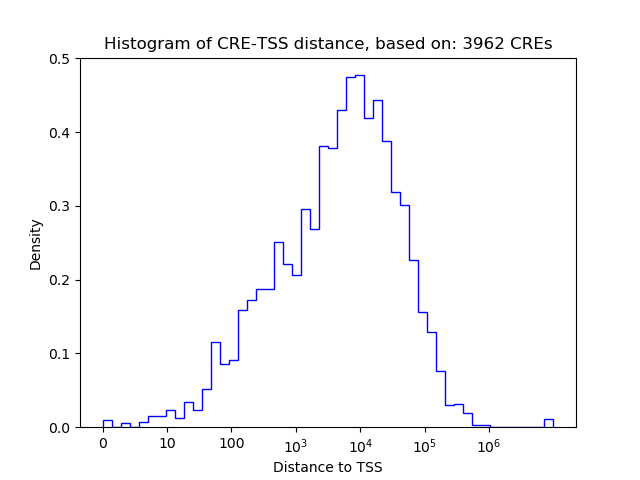
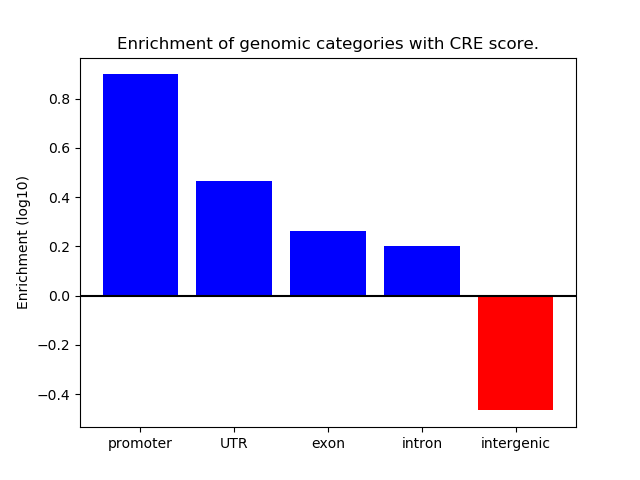
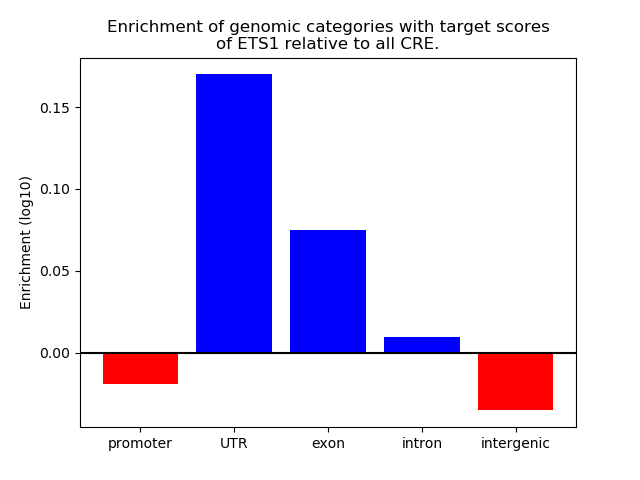
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_127910618_127910769 | ETS1 | 464596 | 0.005531 | 0.81 | 7.8e-03 | Click! |
| chr11_128321217_128321474 | ETS1 | 53944 | 0.155393 | -0.75 | 1.9e-02 | Click! |
| chr11_128089758_128089909 | ETS1 | 285456 | 0.013976 | -0.69 | 4.0e-02 | Click! |
| chr11_128457168_128457319 | ETS1 | 210 | 0.955960 | -0.66 | 5.2e-02 | Click! |
| chr11_128364559_128364710 | ETS1 | 10655 | 0.248798 | 0.64 | 6.5e-02 | Click! |
Activity of the ETS1 motif across conditions
Conditions sorted by the z-value of the ETS1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
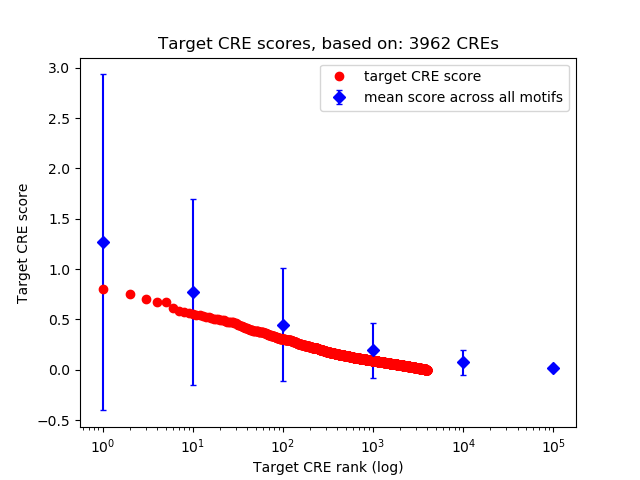
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_242556723_242557374 | 0.80 |
THAP4 |
THAP domain containing 4 |
132 |
0.94 |
| chr22_33040519_33040909 | 0.76 |
ENSG00000251890 |
. |
9530 |
0.25 |
| chrX_129225814_129226276 | 0.71 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
18291 |
0.19 |
| chrX_4158791_4158942 | 0.67 |
ENSG00000264861 |
. |
342675 |
0.01 |
| chr7_150329208_150329551 | 0.67 |
GIMAP6 |
GTPase, IMAP family member 6 |
55 |
0.98 |
| chr1_206732541_206732911 | 0.61 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
2233 |
0.27 |
| chr17_4618141_4618477 | 0.59 |
ARRB2 |
arrestin, beta 2 |
575 |
0.53 |
| chr1_79111525_79111735 | 0.57 |
IFI44 |
interferon-induced protein 44 |
3886 |
0.26 |
| chr2_61095077_61095367 | 0.56 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
13434 |
0.18 |
| chr22_17700031_17700512 | 0.55 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
5 |
0.98 |
| chr10_75625259_75625477 | 0.54 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
4756 |
0.13 |
| chr22_27067492_27067685 | 0.54 |
CRYBA4 |
crystallin, beta A4 |
49660 |
0.13 |
| chr15_100049921_100050072 | 0.54 |
AC015660.1 |
HCG1993240; Serologically defined breast cancer antigen NY-BR-40; Uncharacterized protein |
11418 |
0.22 |
| chr3_101441272_101441423 | 0.53 |
CEP97 |
centrosomal protein 97kDa |
1422 |
0.34 |
| chr22_37637445_37637596 | 0.53 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
2768 |
0.2 |
| chr9_95726568_95726886 | 0.51 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
484 |
0.84 |
| chr2_198157762_198157913 | 0.51 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
9406 |
0.17 |
| chr14_101034010_101035263 | 0.51 |
BEGAIN |
brain-enriched guanylate kinase-associated |
192 |
0.93 |
| chr8_66863582_66864279 | 0.50 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
69865 |
0.12 |
| chr3_67133876_67134122 | 0.49 |
KBTBD8 |
kelch repeat and BTB (POZ) domain containing 8 |
84650 |
0.11 |
| chr3_197831468_197832400 | 0.49 |
AC073135.3 |
|
5143 |
0.19 |
| chr6_225379_225671 | 0.49 |
DUSP22 |
dual specificity phosphatase 22 |
66105 |
0.15 |
| chr6_41168130_41168571 | 0.48 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
582 |
0.65 |
| chr9_126628860_126629011 | 0.48 |
DENND1A |
DENN/MADD domain containing 1A |
63451 |
0.12 |
| chr17_4336857_4337447 | 0.47 |
SPNS3 |
spinster homolog 3 (Drosophila) |
67 |
0.97 |
| chr10_29528746_29529040 | 0.47 |
LYZL1 |
lysozyme-like 1 |
49097 |
0.18 |
| chr21_42741926_42742138 | 0.47 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
20 |
0.98 |
| chr12_122075303_122075715 | 0.47 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
10788 |
0.17 |
| chr1_9777067_9777879 | 0.47 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
654 |
0.69 |
| chr22_37637600_37637886 | 0.46 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
2545 |
0.22 |
| chr2_112913175_112913326 | 0.46 |
FBLN7 |
fibulin 7 |
4075 |
0.29 |
| chr17_38478388_38479657 | 0.45 |
RARA |
retinoic acid receptor, alpha |
4485 |
0.13 |
| chr10_134260489_134261029 | 0.44 |
C10orf91 |
chromosome 10 open reading frame 91 |
2066 |
0.3 |
| chr1_156124352_156124692 | 0.43 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
360 |
0.78 |
| chr2_234327510_234328202 | 0.43 |
DGKD |
diacylglycerol kinase, delta 130kDa |
31056 |
0.14 |
| chr4_2800484_2801318 | 0.43 |
SH3BP2 |
SH3-domain binding protein 2 |
176 |
0.95 |
| chr22_39148225_39148574 | 0.42 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
2080 |
0.14 |
| chr9_132890415_132890884 | 0.42 |
ENSG00000223188 |
. |
7564 |
0.19 |
| chr1_203734376_203734572 | 0.41 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
169 |
0.94 |
| chr3_140985922_140986285 | 0.41 |
ACPL2 |
acid phosphatase-like 2 |
4647 |
0.28 |
| chr12_56323213_56323608 | 0.41 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
802 |
0.4 |
| chr17_80063977_80064793 | 0.40 |
CCDC57 |
coiled-coil domain containing 57 |
4659 |
0.1 |
| chr14_24906611_24906867 | 0.40 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
3518 |
0.13 |
| chr7_50727890_50728167 | 0.39 |
GRB10 |
growth factor receptor-bound protein 10 |
33401 |
0.21 |
| chr7_97878599_97878750 | 0.39 |
TECPR1 |
tectonin beta-propeller repeat containing 1 |
2565 |
0.27 |
| chr10_126300385_126300576 | 0.39 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
91714 |
0.07 |
| chr15_40746159_40746352 | 0.39 |
RP11-64K12.9 |
|
583 |
0.57 |
| chr1_198648918_198649184 | 0.39 |
RP11-553K8.5 |
|
12861 |
0.25 |
| chr17_72459969_72460120 | 0.39 |
CD300A |
CD300a molecule |
2511 |
0.22 |
| chr6_32160504_32160787 | 0.39 |
GPSM3 |
G-protein signaling modulator 3 |
0 |
0.94 |
| chrX_3061320_3061471 | 0.38 |
ARSF |
arylsulfatase F |
76521 |
0.09 |
| chr2_196444875_196445026 | 0.38 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
4249 |
0.31 |
| chr5_172272383_172272534 | 0.38 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
11044 |
0.19 |
| chr8_130983839_130984300 | 0.38 |
FAM49B |
family with sequence similarity 49, member B |
833 |
0.63 |
| chr4_6912691_6912996 | 0.38 |
TBC1D14 |
TBC1 domain family, member 14 |
868 |
0.61 |
| chr3_111264316_111264467 | 0.38 |
CD96 |
CD96 molecule |
3394 |
0.32 |
| chr13_41579755_41579906 | 0.38 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
13620 |
0.19 |
| chr17_11683304_11683455 | 0.37 |
ENSG00000263623 |
. |
38598 |
0.18 |
| chr1_247570490_247571007 | 0.37 |
NLRP3 |
NLR family, pyrin domain containing 3 |
8710 |
0.18 |
| chr5_14594534_14594878 | 0.37 |
FAM105A |
family with sequence similarity 105, member A |
12822 |
0.24 |
| chr9_132023537_132023688 | 0.37 |
ENSG00000220992 |
. |
30616 |
0.15 |
| chr4_140481827_140481978 | 0.37 |
RP11-342I1.2 |
|
3949 |
0.22 |
| chr3_16883969_16884348 | 0.36 |
PLCL2 |
phospholipase C-like 2 |
42294 |
0.19 |
| chr18_74836287_74836734 | 0.36 |
MBP |
myelin basic protein |
3158 |
0.38 |
| chr18_77166229_77166515 | 0.36 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
5980 |
0.26 |
| chr6_35338140_35338402 | 0.36 |
PPARD |
peroxisome proliferator-activated receptor delta |
27869 |
0.16 |
| chr10_119125041_119125192 | 0.36 |
ENSG00000222197 |
. |
7249 |
0.22 |
| chr15_38852692_38852843 | 0.35 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
287 |
0.91 |
| chr10_43947307_43947516 | 0.35 |
ENSG00000252532 |
. |
6102 |
0.17 |
| chr16_28506747_28507012 | 0.35 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
24 |
0.94 |
| chr1_234980313_234980644 | 0.35 |
ENSG00000201638 |
. |
6758 |
0.28 |
| chr16_71915132_71915582 | 0.34 |
RP11-498D10.3 |
|
819 |
0.43 |
| chr3_177662001_177662152 | 0.34 |
ENSG00000199858 |
. |
63491 |
0.15 |
| chr5_137416910_137417200 | 0.34 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
2526 |
0.17 |
| chr2_113873876_113874027 | 0.34 |
IL1RN |
interleukin 1 receptor antagonist |
1519 |
0.32 |
| chr9_95716277_95716712 | 0.34 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
6761 |
0.23 |
| chr12_58158605_58159703 | 0.33 |
CYP27B1 |
cytochrome P450, family 27, subfamily B, polypeptide 1 |
314 |
0.7 |
| chr5_118676288_118676442 | 0.33 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
7495 |
0.22 |
| chr22_50319474_50319638 | 0.33 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
7175 |
0.17 |
| chr6_99792225_99792376 | 0.33 |
FAXC |
failed axon connections homolog (Drosophila) |
1490 |
0.45 |
| chr6_35465033_35465928 | 0.33 |
TEAD3 |
TEA domain family member 3 |
627 |
0.7 |
| chr12_53024457_53024608 | 0.32 |
KRT73 |
keratin 73 |
12189 |
0.1 |
| chr10_73531555_73531706 | 0.32 |
C10orf54 |
chromosome 10 open reading frame 54 |
1625 |
0.38 |
| chr4_38611399_38611550 | 0.32 |
RP11-617D20.1 |
|
14722 |
0.22 |
| chr5_544016_544540 | 0.32 |
ENSG00000264233 |
. |
8281 |
0.14 |
| chr17_47765494_47765708 | 0.32 |
SPOP |
speckle-type POZ protein |
10005 |
0.14 |
| chr12_6571175_6571326 | 0.32 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
8561 |
0.09 |
| chr2_127942699_127942861 | 0.32 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
20563 |
0.22 |
| chr20_23063904_23064055 | 0.32 |
CD93 |
CD93 molecule |
2998 |
0.23 |
| chr15_67442847_67442998 | 0.31 |
RP11-342M21.2 |
Uncharacterized protein |
3645 |
0.27 |
| chr5_176784624_176784924 | 0.31 |
RGS14 |
regulator of G-protein signaling 14 |
64 |
0.95 |
| chr1_25252115_25252332 | 0.31 |
RUNX3 |
runt-related transcription factor 3 |
3389 |
0.28 |
| chr16_79123602_79124194 | 0.31 |
RP11-556H2.3 |
|
49 |
0.98 |
| chr3_15332172_15332523 | 0.31 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
14795 |
0.14 |
| chr3_113252540_113252793 | 0.31 |
SIDT1 |
SID1 transmembrane family, member 1 |
1448 |
0.4 |
| chr7_106505707_106506635 | 0.31 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
247 |
0.95 |
| chr12_54866933_54867431 | 0.31 |
GTSF1 |
gametocyte specific factor 1 |
201 |
0.92 |
| chr17_3693803_3693954 | 0.31 |
ITGAE |
integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
10659 |
0.14 |
| chr16_30915019_30915170 | 0.30 |
CTF1 |
cardiotrophin 1 |
7135 |
0.08 |
| chr6_144665292_144666004 | 0.30 |
UTRN |
utrophin |
411 |
0.88 |
| chr2_101171356_101171507 | 0.30 |
ENSG00000266005 |
. |
1081 |
0.48 |
| chr10_65001333_65001484 | 0.30 |
JMJD1C |
jumonji domain containing 1C |
27418 |
0.22 |
| chr20_35258352_35258894 | 0.30 |
SLA2 |
Src-like-adaptor 2 |
15661 |
0.12 |
| chr22_39147860_39148011 | 0.30 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
1616 |
0.18 |
| chr7_2684869_2685165 | 0.30 |
TTYH3 |
tweety family member 3 |
149 |
0.96 |
| chr11_128587302_128587453 | 0.30 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
21459 |
0.17 |
| chr6_35279100_35279566 | 0.30 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1818 |
0.34 |
| chr14_22979723_22979978 | 0.30 |
TRAJ15 |
T cell receptor alpha joining 15 |
18730 |
0.09 |
| chrX_65294918_65295209 | 0.30 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
35096 |
0.18 |
| chr10_94590680_94591107 | 0.30 |
EXOC6 |
exocyst complex component 6 |
42 |
0.99 |
| chr16_78776918_78777171 | 0.30 |
RP11-319G9.3 |
|
154018 |
0.04 |
| chr17_66344269_66344555 | 0.30 |
ARSG |
arylsulfatase G |
56753 |
0.1 |
| chr11_65407442_65407916 | 0.30 |
SIPA1 |
signal-induced proliferation-associated 1 |
87 |
0.92 |
| chr1_27945148_27945299 | 0.30 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
5350 |
0.15 |
| chr1_90312219_90312370 | 0.30 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
155 |
0.97 |
| chr12_51714745_51715007 | 0.30 |
BIN2 |
bridging integrator 2 |
3023 |
0.21 |
| chr6_36923610_36923761 | 0.29 |
PI16 |
peptidase inhibitor 16 |
1476 |
0.38 |
| chr6_114130917_114131144 | 0.29 |
ENSG00000253091 |
. |
28363 |
0.16 |
| chr1_154925966_154926117 | 0.29 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
2539 |
0.12 |
| chr3_42543385_42544225 | 0.29 |
VIPR1 |
vasoactive intestinal peptide receptor 1 |
299 |
0.89 |
| chr2_12267962_12268113 | 0.29 |
ENSG00000264089 |
. |
71219 |
0.12 |
| chr22_36730567_36730805 | 0.29 |
ENSG00000266345 |
. |
18440 |
0.17 |
| chr14_91882732_91883366 | 0.29 |
CCDC88C |
coiled-coil domain containing 88C |
641 |
0.78 |
| chr22_17566954_17567330 | 0.29 |
IL17RA |
interleukin 17 receptor A |
1293 |
0.41 |
| chr3_188077055_188077206 | 0.29 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
119478 |
0.06 |
| chr22_32039117_32039268 | 0.29 |
PISD |
phosphatidylserine decarboxylase |
4761 |
0.23 |
| chr10_126339527_126339678 | 0.28 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
52592 |
0.13 |
| chr15_65184402_65184553 | 0.28 |
ENSG00000264929 |
. |
7961 |
0.15 |
| chr6_154571300_154571451 | 0.28 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
2559 |
0.43 |
| chr1_2347968_2348198 | 0.28 |
PEX10 |
peroxisomal biogenesis factor 10 |
2847 |
0.15 |
| chr13_30528667_30528971 | 0.28 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
28031 |
0.25 |
| chr2_106363218_106363369 | 0.28 |
NCK2 |
NCK adaptor protein 2 |
1105 |
0.66 |
| chr4_37759806_37759957 | 0.28 |
ENSG00000207075 |
. |
58257 |
0.12 |
| chr14_91608686_91608837 | 0.28 |
C14orf159 |
chromosome 14 open reading frame 159 |
5361 |
0.17 |
| chr7_149647429_149647580 | 0.28 |
ATP6V0E2-AS1 |
ATP6V0E2 antisense RNA 1 |
69805 |
0.1 |
| chr1_212968265_212968416 | 0.28 |
TATDN3 |
TatD DNase domain containing 3 |
315 |
0.88 |
| chr17_62705322_62705475 | 0.27 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
47212 |
0.12 |
| chr1_27286933_27287204 | 0.27 |
C1orf172 |
chromosome 1 open reading frame 172 |
171 |
0.93 |
| chr14_22989819_22990019 | 0.27 |
TRAJ15 |
T cell receptor alpha joining 15 |
8661 |
0.11 |
| chr10_121277005_121277156 | 0.27 |
RGS10 |
regulator of G-protein signaling 10 |
9909 |
0.25 |
| chr19_16698320_16698560 | 0.27 |
MED26 |
mediator complex subunit 26 |
28 |
0.83 |
| chr22_38023673_38023836 | 0.27 |
SH3BP1 |
SH3-domain binding protein 1 |
11728 |
0.1 |
| chr2_51231032_51231183 | 0.27 |
NRXN1 |
neurexin 1 |
24304 |
0.21 |
| chr11_46367528_46367907 | 0.26 |
DGKZ |
diacylglycerol kinase, zeta |
730 |
0.6 |
| chr2_121363693_121363844 | 0.26 |
ENSG00000201006 |
. |
45081 |
0.19 |
| chr10_11187446_11187644 | 0.26 |
CELF2 |
CUGBP, Elav-like family member 2 |
19448 |
0.19 |
| chr12_80747054_80747205 | 0.26 |
OTOGL |
otogelin-like |
3005 |
0.29 |
| chr13_24798397_24798604 | 0.26 |
SPATA13 |
spermatogenesis associated 13 |
27332 |
0.16 |
| chr13_40466947_40467177 | 0.26 |
ENSG00000212553 |
. |
35698 |
0.22 |
| chr17_37033575_37033782 | 0.26 |
LASP1 |
LIM and SH3 protein 1 |
3535 |
0.12 |
| chr1_31222830_31222981 | 0.26 |
LAPTM5 |
lysosomal protein transmembrane 5 |
7762 |
0.16 |
| chr19_35740232_35740383 | 0.26 |
LSR |
lipolysis stimulated lipoprotein receptor |
303 |
0.79 |
| chr10_14659345_14659496 | 0.26 |
FAM107B |
family with sequence similarity 107, member B |
4869 |
0.24 |
| chrX_70838155_70838405 | 0.25 |
CXCR3 |
chemokine (C-X-C motif) receptor 3 |
80 |
0.97 |
| chr12_10220623_10220774 | 0.25 |
ENSG00000223042 |
. |
16087 |
0.12 |
| chrX_56791675_56792116 | 0.25 |
ENSG00000204272 |
. |
36203 |
0.23 |
| chr9_35649649_35650491 | 0.25 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
867 |
0.32 |
| chr12_46611822_46611973 | 0.25 |
SLC38A1 |
solute carrier family 38, member 1 |
49587 |
0.18 |
| chr3_156360650_156360801 | 0.25 |
TIPARP-AS1 |
TIPARP antisense RNA 1 |
29934 |
0.18 |
| chr1_110416375_110416526 | 0.25 |
RP11-195M16.1 |
|
12436 |
0.18 |
| chr1_42073785_42073936 | 0.25 |
RP11-486B10.3 |
|
72746 |
0.1 |
| chr1_154982220_154982994 | 0.25 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
4317 |
0.08 |
| chr5_169075790_169075941 | 0.25 |
DOCK2 |
dedicator of cytokinesis 2 |
11614 |
0.25 |
| chr4_48139910_48140061 | 0.25 |
TXK |
TXK tyrosine kinase |
3712 |
0.2 |
| chr2_175357999_175358525 | 0.25 |
GPR155 |
G protein-coupled receptor 155 |
6440 |
0.21 |
| chr7_26333293_26333815 | 0.25 |
SNX10 |
sorting nexin 10 |
880 |
0.67 |
| chr2_112940980_112941186 | 0.25 |
FBLN7 |
fibulin 7 |
1623 |
0.45 |
| chr1_94459122_94459273 | 0.25 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
17984 |
0.22 |
| chr6_42743118_42743455 | 0.24 |
GLTSCR1L |
GLTSCR1-like |
6479 |
0.18 |
| chr3_197825673_197826397 | 0.24 |
AC073135.3 |
|
11042 |
0.16 |
| chr11_8111770_8111921 | 0.24 |
TUB |
tubby bipartite transcription factor |
8936 |
0.16 |
| chr9_95716734_95716885 | 0.24 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
7076 |
0.22 |
| chr16_28505762_28506015 | 0.24 |
APOBR |
apolipoprotein B receptor |
82 |
0.93 |
| chr14_22343272_22343423 | 0.24 |
ENSG00000222776 |
. |
94562 |
0.08 |
| chr3_183273606_183273960 | 0.24 |
KLHL6 |
kelch-like family member 6 |
306 |
0.89 |
| chr6_143177934_143178085 | 0.24 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
19825 |
0.26 |
| chr19_55084758_55085279 | 0.24 |
LILRA2 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
200 |
0.63 |
| chr10_75624935_75625135 | 0.24 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
4423 |
0.13 |
| chr11_6251374_6251525 | 0.24 |
FAM160A2 |
family with sequence similarity 160, member A2 |
4397 |
0.13 |
| chr15_28381443_28381594 | 0.24 |
OCA2 |
oculocutaneous albinism II |
37014 |
0.18 |
| chr17_25793367_25793654 | 0.24 |
RP11-720N19.2 |
|
1372 |
0.44 |
| chr7_100484820_100485063 | 0.24 |
SRRT |
serrate RNA effector molecule homolog (Arabidopsis) |
383 |
0.71 |
| chr21_15922641_15922820 | 0.24 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
4066 |
0.28 |
| chr8_22437861_22438169 | 0.24 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
12 |
0.95 |
| chr12_104871725_104872091 | 0.24 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
21129 |
0.25 |
| chr7_142000915_142001066 | 0.24 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
11381 |
0.22 |
| chr11_116722246_116722635 | 0.24 |
SIK3 |
SIK family kinase 3 |
2586 |
0.16 |
| chr14_105953334_105954468 | 0.24 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
647 |
0.53 |
| chr8_134084504_134084857 | 0.23 |
SLA |
Src-like-adaptor |
12077 |
0.24 |
| chr22_51059908_51060313 | 0.23 |
ARSA |
arylsulfatase A |
6473 |
0.1 |
| chr6_112078758_112079082 | 0.23 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1397 |
0.55 |
| chr20_8134073_8134224 | 0.23 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
20846 |
0.25 |
| chr4_164480063_164480214 | 0.23 |
ENSG00000264535 |
. |
35694 |
0.17 |
| chr4_125821414_125821565 | 0.23 |
ANKRD50 |
ankyrin repeat domain 50 |
187602 |
0.03 |
| chr3_146520537_146520688 | 0.23 |
ENSG00000251800 |
. |
151253 |
0.04 |
| chrX_131121415_131121857 | 0.23 |
ENSG00000265686 |
. |
9237 |
0.22 |
| chr22_23493647_23493798 | 0.23 |
RAB36 |
RAB36, member RAS oncogene family |
1422 |
0.3 |
| chr6_166747166_166747317 | 0.23 |
SFT2D1 |
SFT2 domain containing 1 |
8838 |
0.19 |
| chr1_161038546_161039545 | 0.23 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr2_237780403_237780554 | 0.23 |
ENSG00000202341 |
. |
143470 |
0.04 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.2 | 0.5 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.4 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.1 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.2 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.9 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 0.2 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.4 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.2 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.2 | GO:0050706 | regulation of interleukin-1 beta secretion(GO:0050706) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.0 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.7 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.0 | GO:0070828 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.4 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 3.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |