Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
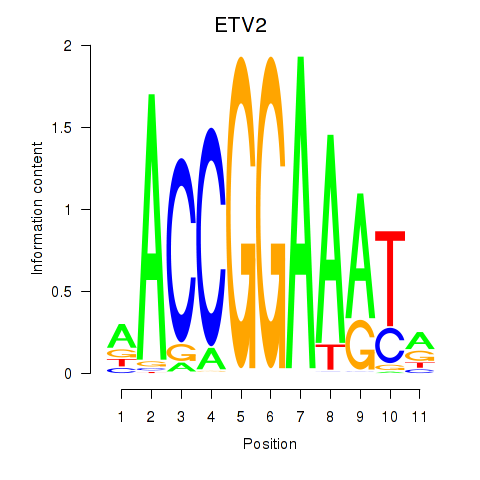
Results for ETV2
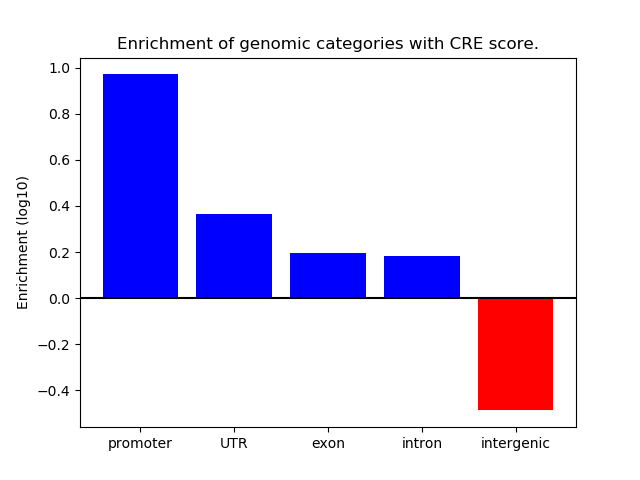
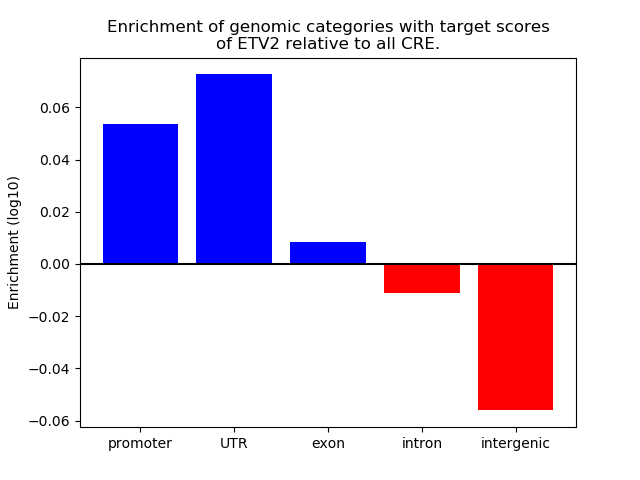
Z-value: 0.96
Transcription factors associated with ETV2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV2
|
ENSG00000105672.10 | ETS variant transcription factor 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_36136140_36136291 | ETV2 | 1647 | 0.171867 | -0.35 | 3.6e-01 | Click! |
| chr19_36132782_36132933 | ETV2 | 162 | 0.884234 | 0.12 | 7.7e-01 | Click! |
| chr19_36132415_36132566 | ETV2 | 157 | 0.887775 | 0.09 | 8.3e-01 | Click! |
| chr19_36135542_36136001 | ETV2 | 1203 | 0.243740 | 0.08 | 8.4e-01 | Click! |
| chr19_36134803_36135339 | ETV2 | 503 | 0.581797 | -0.07 | 8.6e-01 | Click! |
Activity of the ETV2 motif across conditions
Conditions sorted by the z-value of the ETV2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
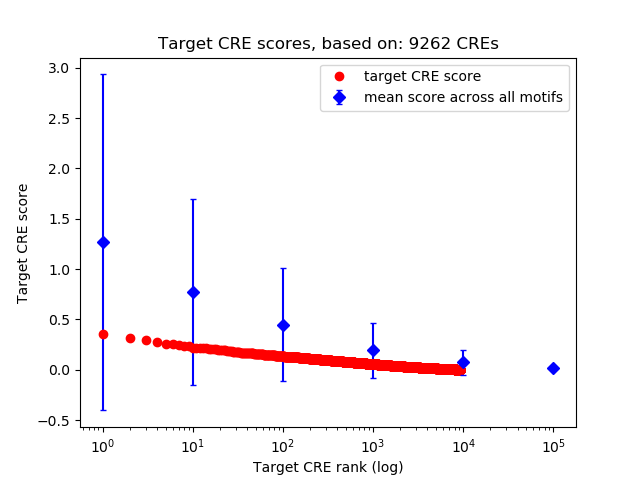
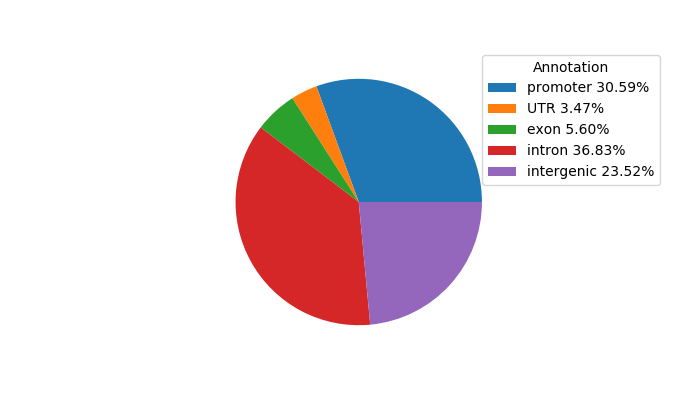
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_13464907_13465379 | 0.35 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
129 |
0.93 |
| chrX_111125456_111125607 | 0.31 |
TRPC5OS |
TRPC5 opposite strand |
406 |
0.91 |
| chr5_128680466_128680617 | 0.29 |
ENSG00000265691 |
. |
52299 |
0.16 |
| chr7_150180608_150181504 | 0.27 |
GIMAP7 |
GTPase, IMAP family member 7 |
30862 |
0.13 |
| chr10_45409178_45409329 | 0.26 |
TMEM72 |
transmembrane protein 72 |
2489 |
0.23 |
| chr16_30915019_30915170 | 0.25 |
CTF1 |
cardiotrophin 1 |
7135 |
0.08 |
| chr5_57788132_57788386 | 0.25 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
995 |
0.57 |
| chr16_89520011_89520290 | 0.23 |
ENSG00000252887 |
. |
10424 |
0.12 |
| chr1_239883177_239883989 | 0.23 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
740 |
0.59 |
| chr12_11954842_11955089 | 0.22 |
ETV6 |
ets variant 6 |
49530 |
0.18 |
| chr4_118904778_118905073 | 0.22 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
50575 |
0.19 |
| chr7_150329208_150329551 | 0.21 |
GIMAP6 |
GTPase, IMAP family member 6 |
55 |
0.98 |
| chr12_104871725_104872091 | 0.21 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
21129 |
0.25 |
| chr11_117873994_117874145 | 0.21 |
IL10RA |
interleukin 10 receptor, alpha |
16960 |
0.17 |
| chr12_9912315_9912520 | 0.21 |
CD69 |
CD69 molecule |
1080 |
0.47 |
| chr11_65409143_65409294 | 0.21 |
SIPA1 |
signal-induced proliferation-associated 1 |
919 |
0.33 |
| chr9_92092302_92093120 | 0.20 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
2094 |
0.37 |
| chr2_105461225_105461520 | 0.20 |
LINC01158 |
long intergenic non-protein coding RNA 1158 |
6539 |
0.15 |
| chr12_51640112_51640647 | 0.20 |
DAZAP2 |
DAZ associated protein 2 |
7300 |
0.14 |
| chr4_1717124_1717335 | 0.20 |
SLBP |
stem-loop binding protein |
2947 |
0.18 |
| chr9_78286476_78286627 | 0.20 |
ENSG00000238598 |
. |
5272 |
0.3 |
| chr19_38539731_38539968 | 0.19 |
ENSG00000221258 |
. |
4756 |
0.2 |
| chr17_56415064_56415366 | 0.19 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
496 |
0.64 |
| chrX_111125134_111125285 | 0.19 |
TRPC5OS |
TRPC5 opposite strand |
84 |
0.98 |
| chr9_71611525_71611676 | 0.19 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
17439 |
0.21 |
| chr6_139646858_139647120 | 0.18 |
TXLNB |
taxilin beta |
33713 |
0.16 |
| chr19_44283473_44283624 | 0.18 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
1861 |
0.24 |
| chr2_230989553_230989704 | 0.18 |
AC009950.2 |
|
398 |
0.87 |
| chr1_161038546_161039545 | 0.18 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr3_128211427_128211634 | 0.18 |
GATA2 |
GATA binding protein 2 |
486 |
0.76 |
| chr13_49828247_49828398 | 0.17 |
CDADC1 |
cytidine and dCMP deaminase domain containing 1 |
6138 |
0.24 |
| chr1_158624437_158624588 | 0.17 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
31886 |
0.12 |
| chr1_221246452_221246603 | 0.17 |
HLX |
H2.0-like homeobox |
191943 |
0.03 |
| chr5_59544424_59544722 | 0.17 |
ENSG00000202017 |
. |
55405 |
0.15 |
| chr10_15356030_15356181 | 0.17 |
FAM171A1 |
family with sequence similarity 171, member A1 |
56953 |
0.14 |
| chr1_66802739_66802890 | 0.17 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
4942 |
0.34 |
| chr6_119764039_119764190 | 0.17 |
MAN1A1 |
mannosidase, alpha, class 1A, member 1 |
93188 |
0.09 |
| chr15_32754386_32754537 | 0.17 |
GOLGA8O |
golgin A8 family, member O |
6626 |
0.11 |
| chr21_45577511_45577678 | 0.17 |
AP001055.1 |
|
15986 |
0.13 |
| chr7_150374007_150374169 | 0.17 |
GIMAP2 |
GTPase, IMAP family member 2 |
8700 |
0.17 |
| chr12_10396486_10396819 | 0.17 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
17995 |
0.13 |
| chr21_16433048_16433989 | 0.17 |
NRIP1 |
nuclear receptor interacting protein 1 |
3608 |
0.34 |
| chr7_38352490_38352648 | 0.17 |
STARD3NL |
STARD3 N-terminal like |
134572 |
0.05 |
| chr18_20254692_20254843 | 0.17 |
ENSG00000206827 |
. |
27222 |
0.21 |
| chr20_3888044_3888195 | 0.16 |
ENSG00000199024 |
. |
10022 |
0.14 |
| chr10_6604051_6604202 | 0.16 |
PRKCQ |
protein kinase C, theta |
18075 |
0.29 |
| chr12_92937765_92937916 | 0.16 |
ENSG00000238865 |
. |
923 |
0.69 |
| chr12_15134766_15134917 | 0.16 |
PDE6H |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
8885 |
0.18 |
| chr2_44464528_44464816 | 0.16 |
ENSG00000239052 |
. |
2128 |
0.31 |
| chr20_43525395_43525569 | 0.16 |
YWHAB |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
11089 |
0.14 |
| chr2_68966105_68966288 | 0.16 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4182 |
0.3 |
| chr2_106776864_106777181 | 0.16 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
18 |
0.98 |
| chr3_47059139_47059290 | 0.16 |
NBEAL2 |
neurobeachin-like 2 |
12443 |
0.18 |
| chr2_234406495_234406646 | 0.16 |
USP40 |
ubiquitin specific peptidase 40 |
8463 |
0.22 |
| chr16_24994986_24995137 | 0.16 |
ARHGAP17 |
Rho GTPase activating protein 17 |
31591 |
0.21 |
| chr12_72039123_72039274 | 0.16 |
ENSG00000212461 |
. |
6063 |
0.17 |
| chr19_52274682_52274833 | 0.15 |
FPR2 |
formyl peptide receptor 2 |
3126 |
0.17 |
| chr12_8309713_8309864 | 0.15 |
ZNF705A |
zinc finger protein 705A |
158 |
0.94 |
| chrX_138636602_138636753 | 0.15 |
F9 |
coagulation factor IX |
23753 |
0.2 |
| chr5_130715501_130715652 | 0.15 |
CDC42SE2 |
CDC42 small effector 2 |
5723 |
0.32 |
| chr8_144437885_144438036 | 0.15 |
TOP1MT |
topoisomerase (DNA) I, mitochondrial |
256 |
0.84 |
| chr9_135248236_135248390 | 0.15 |
SETX |
senataxin |
17941 |
0.17 |
| chr2_175354938_175355089 | 0.15 |
GPR155 |
G protein-coupled receptor 155 |
3191 |
0.26 |
| chr11_14600078_14600336 | 0.15 |
PSMA1 |
proteasome (prosome, macropain) subunit, alpha type, 1 |
57965 |
0.12 |
| chr2_68592484_68592752 | 0.15 |
AC015969.3 |
|
98 |
0.77 |
| chr21_36772376_36772631 | 0.15 |
ENSG00000211590 |
. |
320510 |
0.01 |
| chr12_2173143_2173560 | 0.15 |
CACNA1C |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
10622 |
0.17 |
| chr7_148001198_148001349 | 0.15 |
ENSG00000199370 |
. |
85468 |
0.1 |
| chr2_231733015_231733166 | 0.15 |
ITM2C |
integral membrane protein 2C |
2711 |
0.25 |
| chr5_74964815_74965486 | 0.15 |
ENSG00000207333 |
. |
40274 |
0.14 |
| chr15_50570751_50570902 | 0.15 |
HDC |
histidine decarboxylase |
12603 |
0.14 |
| chr7_2286127_2286278 | 0.15 |
NUDT1 |
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
3662 |
0.2 |
| chr17_73064038_73064189 | 0.15 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
19709 |
0.07 |
| chr13_91922419_91922570 | 0.15 |
ENSG00000215417 |
. |
80365 |
0.12 |
| chr21_15892777_15892928 | 0.15 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
25810 |
0.22 |
| chr1_22402292_22402667 | 0.15 |
CDC42-IT1 |
CDC42 intronic transcript 1 (non-protein coding) |
16789 |
0.13 |
| chr13_52570204_52570355 | 0.14 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
15268 |
0.17 |
| chr14_25143618_25143855 | 0.14 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
40263 |
0.14 |
| chr10_129861236_129861473 | 0.14 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
15520 |
0.26 |
| chr2_233703428_233703579 | 0.14 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
4167 |
0.17 |
| chr6_53187554_53187810 | 0.14 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
25905 |
0.17 |
| chr4_16600596_16600747 | 0.14 |
LDB2 |
LIM domain binding 2 |
3173 |
0.4 |
| chr9_96929356_96930067 | 0.14 |
ENSG00000199165 |
. |
8523 |
0.16 |
| chr7_93485635_93485786 | 0.14 |
TFPI2 |
tissue factor pathway inhibitor 2 |
33772 |
0.15 |
| chr7_79736330_79736481 | 0.14 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
26866 |
0.25 |
| chr12_68953962_68954113 | 0.14 |
RAP1B |
RAP1B, member of RAS oncogene family |
50582 |
0.15 |
| chr8_130984307_130984565 | 0.14 |
FAM49B |
family with sequence similarity 49, member B |
1200 |
0.49 |
| chr14_61966690_61966841 | 0.14 |
RP11-47I22.4 |
|
29081 |
0.17 |
| chr6_15439743_15439894 | 0.14 |
JARID2 |
jumonji, AT rich interactive domain 2 |
38729 |
0.19 |
| chr15_101675976_101676127 | 0.14 |
RP11-505E24.2 |
|
49780 |
0.14 |
| chr12_27166173_27166337 | 0.14 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
308 |
0.87 |
| chr2_54816819_54816970 | 0.14 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
31363 |
0.16 |
| chr7_4901206_4901513 | 0.14 |
PAPOLB |
poly(A) polymerase beta (testis specific) |
266 |
0.91 |
| chr11_1779321_1779700 | 0.14 |
RP11-295K3.1 |
|
771 |
0.4 |
| chr13_74339708_74339938 | 0.14 |
KLF12 |
Kruppel-like factor 12 |
229363 |
0.02 |
| chr15_34620156_34620427 | 0.14 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
8754 |
0.11 |
| chr10_94581983_94582410 | 0.14 |
EXOC6 |
exocyst complex component 6 |
8739 |
0.28 |
| chr14_70169921_70170072 | 0.14 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
23621 |
0.21 |
| chr22_17956680_17956831 | 0.13 |
CECR2 |
cat eye syndrome chromosome region, candidate 2 |
128 |
0.97 |
| chr17_77755824_77755975 | 0.13 |
CBX2 |
chromobox homolog 2 |
3906 |
0.16 |
| chr19_55518051_55518213 | 0.13 |
CTC-550B14.7 |
|
559 |
0.64 |
| chr6_108775089_108775240 | 0.13 |
ENSG00000200799 |
. |
61887 |
0.13 |
| chr10_126753392_126753897 | 0.13 |
ENSG00000264572 |
. |
32205 |
0.18 |
| chr22_17700031_17700512 | 0.13 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
5 |
0.98 |
| chr15_68967503_68967698 | 0.13 |
CORO2B |
coronin, actin binding protein, 2B |
43273 |
0.18 |
| chr22_39339167_39339454 | 0.13 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
9446 |
0.14 |
| chr17_76912321_76912483 | 0.13 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
6081 |
0.15 |
| chr12_14410123_14410274 | 0.13 |
ENSG00000251908 |
. |
6923 |
0.23 |
| chr5_126323849_126324159 | 0.13 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
42496 |
0.18 |
| chr15_66906109_66906400 | 0.13 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
31726 |
0.13 |
| chr19_3136242_3137376 | 0.13 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
618 |
0.58 |
| chr19_3812381_3812532 | 0.13 |
MATK |
megakaryocyte-associated tyrosine kinase |
10329 |
0.11 |
| chr15_50411143_50411548 | 0.13 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
74 |
0.98 |
| chr6_170404507_170404739 | 0.13 |
RP11-302L19.1 |
|
73118 |
0.11 |
| chr15_64231749_64232239 | 0.13 |
RP11-111E14.1 |
|
11521 |
0.22 |
| chr11_78061880_78062257 | 0.13 |
GAB2 |
GRB2-associated binding protein 2 |
9142 |
0.19 |
| chr8_8648717_8648916 | 0.13 |
CLDN23 |
claudin 23 |
89368 |
0.08 |
| chr12_111883856_111884007 | 0.13 |
ATXN2 |
ataxin 2 |
9985 |
0.18 |
| chr10_7513829_7514374 | 0.13 |
ENSG00000207453 |
. |
14670 |
0.24 |
| chr17_7239932_7240900 | 0.13 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
507 |
0.51 |
| chr9_124248312_124248463 | 0.13 |
ENSG00000240299 |
. |
8410 |
0.21 |
| chr7_138779576_138780000 | 0.13 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
14312 |
0.2 |
| chr11_128554784_128555228 | 0.13 |
RP11-744N12.3 |
|
1317 |
0.34 |
| chr7_128777310_128777461 | 0.13 |
TSPAN33 |
tetraspanin 33 |
7327 |
0.16 |
| chr1_240655111_240655262 | 0.13 |
AL646016.1 |
Uncharacterized protein |
11682 |
0.21 |
| chrX_111124795_111124946 | 0.13 |
TRPC5OS |
TRPC5 opposite strand |
255 |
0.96 |
| chr7_137619960_137620169 | 0.13 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
1428 |
0.46 |
| chr19_10450524_10450675 | 0.13 |
ICAM3 |
intercellular adhesion molecule 3 |
100 |
0.92 |
| chr15_52121866_52123371 | 0.13 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
791 |
0.61 |
| chr17_48393570_48393721 | 0.13 |
RP11-893F2.9 |
|
28429 |
0.09 |
| chr10_11252091_11252492 | 0.13 |
RP3-323N1.2 |
|
38952 |
0.17 |
| chr2_136690107_136690258 | 0.13 |
DARS |
aspartyl-tRNA synthetase |
12059 |
0.21 |
| chr20_23065943_23066176 | 0.13 |
CD93 |
CD93 molecule |
918 |
0.56 |
| chrX_9910123_9910274 | 0.13 |
AC002365.1 |
Homo sapiens uncharacterized LOC100288814 (LOC100288814), mRNA. |
25194 |
0.17 |
| chr11_126206552_126206753 | 0.13 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
18903 |
0.11 |
| chr11_3968342_3968493 | 0.13 |
STIM1 |
stromal interaction molecule 1 |
156 |
0.95 |
| chr14_60620645_60620999 | 0.13 |
DHRS7 |
dehydrogenase/reductase (SDR family) member 7 |
930 |
0.61 |
| chr18_43405356_43405507 | 0.12 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
46 |
0.98 |
| chr16_57576997_57577182 | 0.12 |
GPR114 |
G protein-coupled receptor 114 |
488 |
0.75 |
| chr15_101472786_101472937 | 0.12 |
LRRK1 |
leucine-rich repeat kinase 1 |
13311 |
0.19 |
| chr2_46146841_46146992 | 0.12 |
PRKCE |
protein kinase C, epsilon |
81125 |
0.11 |
| chr7_26411915_26412066 | 0.12 |
AC004540.4 |
|
3902 |
0.26 |
| chr9_90132771_90132922 | 0.12 |
DAPK1 |
death-associated protein kinase 1 |
18926 |
0.23 |
| chr19_6100497_6100709 | 0.12 |
CTB-66B24.1 |
|
8730 |
0.14 |
| chr2_225792696_225792947 | 0.12 |
DOCK10 |
dedicator of cytokinesis 10 |
18961 |
0.28 |
| chr13_31270446_31270597 | 0.12 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
39124 |
0.18 |
| chr12_54149865_54150016 | 0.12 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
28411 |
0.13 |
| chr2_42598602_42598753 | 0.12 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
2527 |
0.35 |
| chr5_130882612_130883040 | 0.12 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
14100 |
0.29 |
| chr2_10263974_10264125 | 0.12 |
RRM2 |
ribonucleotide reductase M2 |
751 |
0.54 |
| chr17_1547959_1548110 | 0.12 |
SCARF1 |
scavenger receptor class F, member 1 |
1004 |
0.36 |
| chr15_65162716_65162867 | 0.12 |
AC069368.3 |
Uncharacterized protein |
28644 |
0.11 |
| chr1_8971746_8971897 | 0.12 |
ENSG00000201725 |
. |
28229 |
0.11 |
| chr4_78067033_78067281 | 0.12 |
ENSG00000201641 |
. |
4433 |
0.25 |
| chrX_131155738_131155889 | 0.12 |
MST4 |
Serine/threonine-protein kinase MST4 |
1480 |
0.46 |
| chr5_39270144_39270372 | 0.12 |
FYB |
FYN binding protein |
488 |
0.88 |
| chr11_118085317_118085565 | 0.12 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
165 |
0.94 |
| chr10_7199494_7199645 | 0.12 |
SFMBT2 |
Scm-like with four mbt domains 2 |
251138 |
0.02 |
| chr3_148881073_148881224 | 0.12 |
CP |
ceruloplasmin (ferroxidase) |
16260 |
0.18 |
| chr2_27108927_27109078 | 0.12 |
DPYSL5 |
dihydropyrimidinase-like 5 |
37626 |
0.13 |
| chr10_967380_967531 | 0.12 |
LARP4B |
La ribonucleoprotein domain family, member 4B |
10109 |
0.17 |
| chrX_118815979_118816490 | 0.12 |
SEPT6 |
septin 6 |
10558 |
0.17 |
| chr4_99368056_99368277 | 0.12 |
RP11-724M22.1 |
|
49349 |
0.16 |
| chr5_134761441_134761660 | 0.12 |
C5orf20 |
chromosome 5 open reading frame 20 |
21488 |
0.12 |
| chr17_62159661_62159890 | 0.12 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
47710 |
0.1 |
| chr2_101196975_101197172 | 0.12 |
ENSG00000238328 |
. |
3142 |
0.22 |
| chr22_38023673_38023836 | 0.12 |
SH3BP1 |
SH3-domain binding protein 1 |
11728 |
0.1 |
| chr12_47609896_47610314 | 0.12 |
PCED1B |
PC-esterase domain containing 1B |
53 |
0.98 |
| chr7_23511681_23511832 | 0.12 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
1670 |
0.34 |
| chr6_41168130_41168571 | 0.12 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
582 |
0.65 |
| chr1_93644902_93645466 | 0.12 |
CCDC18 |
coiled-coil domain containing 18 |
292 |
0.74 |
| chr7_15276818_15277037 | 0.12 |
AGMO |
alkylglycerol monooxygenase |
128885 |
0.06 |
| chr12_50911115_50911266 | 0.12 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
12300 |
0.22 |
| chr7_1572516_1572667 | 0.12 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
2241 |
0.23 |
| chr3_28390062_28390213 | 0.12 |
AZI2 |
5-azacytidine induced 2 |
17 |
0.91 |
| chr1_158978812_158979065 | 0.12 |
IFI16 |
interferon, gamma-inducible protein 16 |
153 |
0.96 |
| chr1_182237824_182238059 | 0.12 |
ENSG00000206764 |
. |
58262 |
0.14 |
| chr2_70311208_70311359 | 0.12 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
1422 |
0.35 |
| chr5_158444877_158445028 | 0.12 |
CTD-2363C16.2 |
|
32138 |
0.18 |
| chr6_114192994_114193566 | 0.12 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
14739 |
0.18 |
| chr20_4792501_4792764 | 0.12 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
3137 |
0.27 |
| chr6_110743031_110743182 | 0.12 |
DDO |
D-aspartate oxidase |
6341 |
0.19 |
| chr2_111610333_111610484 | 0.12 |
ACOXL |
acyl-CoA oxidase-like |
47512 |
0.19 |
| chr1_200209996_200210147 | 0.12 |
ENSG00000221403 |
. |
96109 |
0.08 |
| chr8_17601172_17601375 | 0.12 |
ENSG00000212280 |
. |
4354 |
0.2 |
| chr1_33399662_33399813 | 0.11 |
ENSG00000221423 |
. |
7842 |
0.14 |
| chr1_6086479_6086932 | 0.11 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
325 |
0.88 |
| chr8_61816196_61816783 | 0.11 |
RP11-33I11.2 |
|
94324 |
0.09 |
| chr7_26141262_26141558 | 0.11 |
ENSG00000266430 |
. |
41451 |
0.15 |
| chr4_109267462_109267684 | 0.11 |
ENSG00000232021 |
. |
170129 |
0.03 |
| chr14_93540208_93540361 | 0.11 |
ITPK1-AS1 |
ITPK1 antisense RNA 1 |
6487 |
0.21 |
| chr16_80598262_80598561 | 0.11 |
RP11-525K10.3 |
|
1379 |
0.4 |
| chr21_36881466_36881794 | 0.11 |
ENSG00000211590 |
. |
211383 |
0.02 |
| chr16_23864221_23864482 | 0.11 |
PRKCB |
protein kinase C, beta |
15807 |
0.23 |
| chr2_200333840_200333991 | 0.11 |
SATB2-AS1 |
SATB2 antisense RNA 1 |
306 |
0.9 |
| chr2_16084260_16084411 | 0.11 |
MYCNOS |
MYCN opposite strand |
1964 |
0.28 |
| chr15_60296023_60296174 | 0.11 |
FOXB1 |
forkhead box B1 |
323 |
0.94 |
| chr13_78596306_78596606 | 0.11 |
RNF219-AS1 |
RNF219 antisense RNA 1 |
32524 |
0.21 |
| chr15_66702574_66702725 | 0.11 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
23494 |
0.11 |
| chr3_53206269_53206420 | 0.11 |
PRKCD |
protein kinase C, delta |
7199 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.3 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0071220 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0010822 | positive regulation of mitochondrion organization(GO:0010822) positive regulation of release of cytochrome c from mitochondria(GO:0090200) positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.0 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.2 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0032633 | interleukin-4 production(GO:0032633) regulation of interleukin-4 production(GO:0032673) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.1 | GO:0045350 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0032401 | establishment of melanosome localization(GO:0032401) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | Genes involved in Insulin receptor signalling cascade |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.0 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |