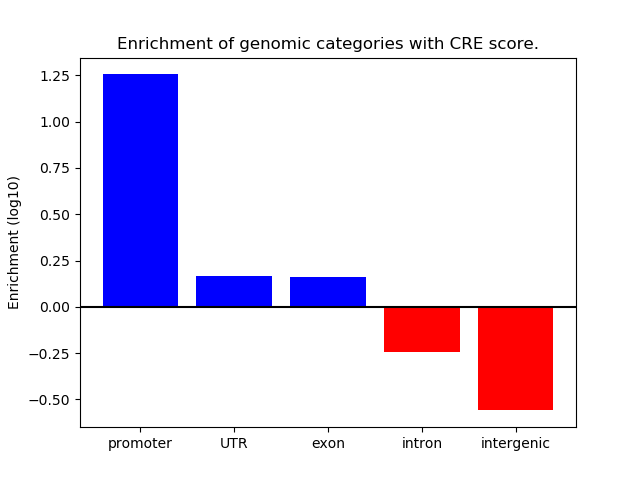
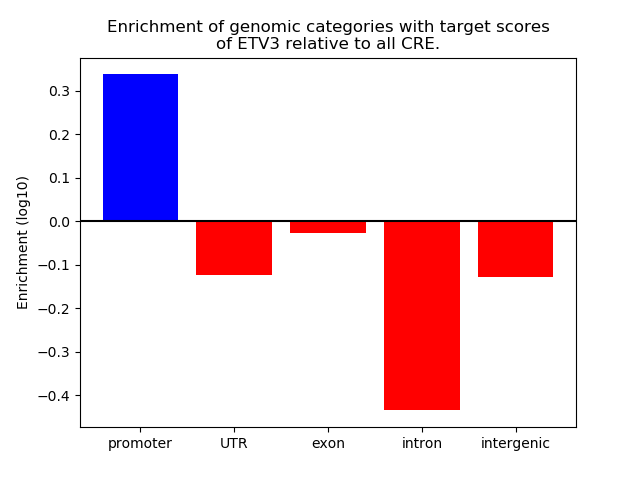
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
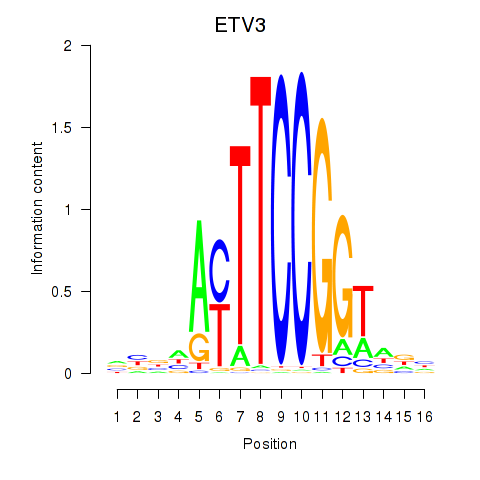
Results for ETV3
Z-value: 0.40
Transcription factors associated with ETV3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV3
|
ENSG00000117036.7 | ETS variant transcription factor 3 |
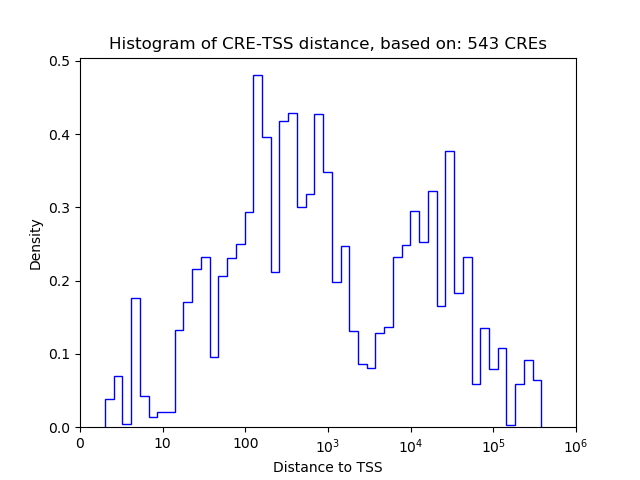
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_157107163_157108171 | ETV3 | 492 | 0.834980 | 0.87 | 2.2e-03 | Click! |
| chr1_157108332_157108851 | ETV3 | 325 | 0.910718 | 0.72 | 2.9e-02 | Click! |
| chr1_157106972_157107150 | ETV3 | 1098 | 0.566512 | 0.57 | 1.1e-01 | Click! |
| chr1_157133708_157133859 | ETV3 | 25517 | 0.204100 | 0.50 | 1.7e-01 | Click! |
| chr1_157114840_157114991 | ETV3 | 6649 | 0.241447 | 0.49 | 1.8e-01 | Click! |
Activity of the ETV3 motif across conditions
Conditions sorted by the z-value of the ETV3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
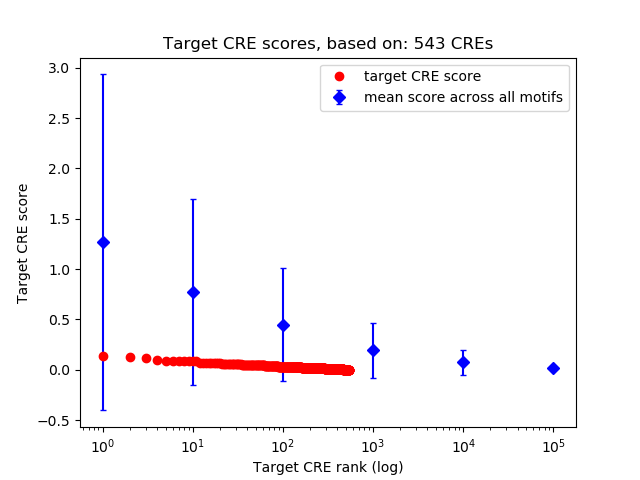
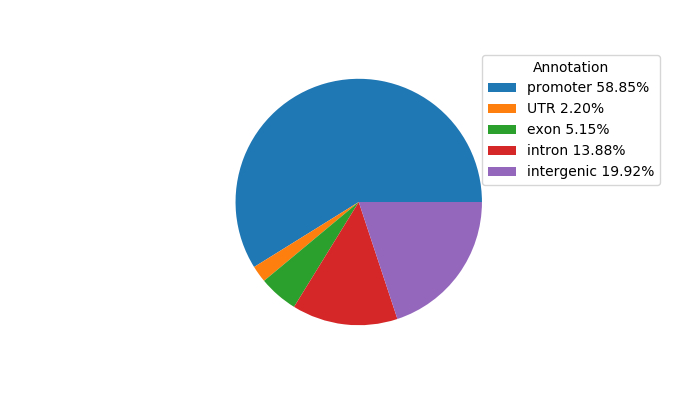
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_52751128_52751393 | 0.14 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
142 |
0.97 |
| chr16_87850031_87850182 | 0.13 |
RP4-536B24.2 |
|
20032 |
0.16 |
| chr6_26026782_26027875 | 0.12 |
HIST1H4B |
histone cluster 1, H4b |
152 |
0.87 |
| chr20_62339371_62339522 | 0.09 |
ZGPAT |
zinc finger, CCCH-type with G patch domain |
4 |
0.53 |
| chr15_101675976_101676127 | 0.09 |
RP11-505E24.2 |
|
49780 |
0.14 |
| chr2_43447013_43447239 | 0.09 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
6622 |
0.24 |
| chr8_121457410_121457992 | 0.09 |
MTBP |
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
35 |
0.54 |
| chr1_224622519_224623397 | 0.09 |
WDR26 |
WD repeat domain 26 |
957 |
0.59 |
| chr5_10250012_10250238 | 0.09 |
CCT5 |
chaperonin containing TCP1, subunit 5 (epsilon) |
84 |
0.53 |
| chrX_86773476_86773748 | 0.08 |
KLHL4 |
kelch-like family member 4 |
795 |
0.74 |
| chr1_54518261_54518829 | 0.08 |
TMEM59 |
transmembrane protein 59 |
320 |
0.67 |
| chr17_13504273_13504480 | 0.07 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
868 |
0.71 |
| chr3_128879854_128880178 | 0.07 |
ISY1 |
ISY1 splicing factor homolog (S. cerevisiae) |
120 |
0.51 |
| chr6_12483851_12484268 | 0.07 |
ENSG00000223321 |
. |
77046 |
0.12 |
| chr10_6836277_6836922 | 0.07 |
PRKCQ |
protein kinase C, theta |
214336 |
0.02 |
| chr18_61088602_61089320 | 0.07 |
VPS4B |
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
662 |
0.7 |
| chr19_16295903_16296067 | 0.06 |
FAM32A |
family with sequence similarity 32, member A |
206 |
0.9 |
| chr16_90113461_90113700 | 0.06 |
URAHP |
urate (hydroxyiso-) hydrolase, pseudogene |
425 |
0.76 |
| chr3_51429228_51429432 | 0.06 |
RBM15B |
RNA binding motif protein 15B |
599 |
0.75 |
| chr4_189329879_189330030 | 0.06 |
RP11-366H4.3 |
|
265421 |
0.02 |
| chr19_55896525_55896972 | 0.06 |
RPL28 |
ribosomal protein L28 |
35 |
0.9 |
| chr16_4186481_4186632 | 0.06 |
ADCY9 |
adenylate cyclase 9 |
20370 |
0.21 |
| chr19_51871127_51871381 | 0.06 |
CLDND2 |
claudin domain containing 2 |
23 |
0.92 |
| chr15_44828103_44829028 | 0.06 |
EIF3J |
eukaryotic translation initiation factor 3, subunit J |
690 |
0.63 |
| chrX_69654840_69655096 | 0.06 |
DLG3 |
discs, large homolog 3 (Drosophila) |
9743 |
0.14 |
| chr11_15096182_15096333 | 0.06 |
CALCB |
calcitonin-related polypeptide beta |
1149 |
0.59 |
| chr4_171011215_171011473 | 0.06 |
AADAT |
aminoadipate aminotransferase |
21 |
0.99 |
| chr19_10443271_10443899 | 0.06 |
RAVER1 |
ribonucleoprotein, PTB-binding 1 |
731 |
0.41 |
| chr12_45626448_45626744 | 0.06 |
ANO6 |
anoctamin 6 |
16693 |
0.28 |
| chr10_121065467_121065729 | 0.06 |
RP11-79M19.2 |
|
30096 |
0.16 |
| chr8_27169019_27169417 | 0.06 |
PTK2B |
protein tyrosine kinase 2 beta |
72 |
0.84 |
| chr19_10305012_10305564 | 0.06 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
14 |
0.96 |
| chr14_78082774_78083096 | 0.05 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
181 |
0.95 |
| chr15_93428286_93428633 | 0.05 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
1933 |
0.32 |
| chr8_41142847_41142998 | 0.05 |
CTD-3080F16.3 |
|
8400 |
0.16 |
| chr2_238875546_238875758 | 0.05 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
4 |
0.98 |
| chr4_26075758_26075909 | 0.05 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
89244 |
0.1 |
| chr11_100557988_100558213 | 0.05 |
ARHGAP42 |
Rho GTPase activating protein 42 |
284 |
0.73 |
| chr16_3481535_3482301 | 0.05 |
ZNF597 |
zinc finger protein 597 |
11624 |
0.11 |
| chr11_44338057_44338887 | 0.05 |
ALX4 |
ALX homeobox 4 |
6756 |
0.31 |
| chr2_24270267_24270418 | 0.05 |
C2orf44 |
chromosome 2 open reading frame 44 |
111 |
0.92 |
| chr17_79195675_79195980 | 0.05 |
AZI1 |
5-azacytidine induced 1 |
903 |
0.39 |
| chr8_28560557_28560810 | 0.05 |
EXTL3-AS1 |
EXTL3 antisense RNA 1 |
1702 |
0.31 |
| chr2_119593693_119594050 | 0.05 |
EN1 |
engrailed homeobox 1 |
11383 |
0.28 |
| chr17_43250076_43250792 | 0.05 |
RP13-890H12.2 |
|
1483 |
0.24 |
| chr8_124693209_124693578 | 0.05 |
KLHL38 |
kelch-like family member 38 |
28203 |
0.16 |
| chr15_77713058_77713524 | 0.05 |
HMG20A |
high mobility group 20A |
28 |
0.95 |
| chr19_55791240_55791709 | 0.05 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
5 |
0.94 |
| chr16_8972178_8972329 | 0.05 |
RP11-77H9.6 |
|
9145 |
0.13 |
| chr10_43249759_43250194 | 0.05 |
BMS1 |
BMS1 ribosome biogenesis factor |
28273 |
0.19 |
| chr1_92952215_92952548 | 0.05 |
GFI1 |
growth factor independent 1 transcription repressor |
52 |
0.98 |
| chr7_74572135_74572677 | 0.05 |
GTF2IRD2B |
GTF2I repeat domain containing 2B |
15502 |
0.24 |
| chr6_153323257_153323895 | 0.05 |
MTRF1L |
mitochondrial translational release factor 1-like |
244 |
0.91 |
| chr13_21287965_21288116 | 0.05 |
ENSG00000265710 |
. |
10198 |
0.19 |
| chr7_154795580_154795843 | 0.05 |
PAXIP1 |
PAX interacting (with transcription-activation domain) protein 1 |
917 |
0.58 |
| chr7_100861322_100861626 | 0.05 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
199 |
0.75 |
| chr12_133613669_133614058 | 0.05 |
ZNF84 |
zinc finger protein 84 |
15 |
0.97 |
| chr17_7387752_7387955 | 0.04 |
POLR2A |
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
86 |
0.71 |
| chr12_42877069_42877413 | 0.04 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
175 |
0.94 |
| chrX_152200749_152200932 | 0.04 |
PNMA3 |
paraneoplastic Ma antigen 3 |
23926 |
0.12 |
| chr1_233249017_233249707 | 0.04 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
46691 |
0.19 |
| chr2_11989597_11989775 | 0.04 |
ENSG00000265172 |
. |
12574 |
0.22 |
| chr1_247275087_247275577 | 0.04 |
C1orf229 |
chromosome 1 open reading frame 229 |
387 |
0.83 |
| chr17_71301523_71301822 | 0.04 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
5875 |
0.2 |
| chr1_19407511_19408053 | 0.04 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
952 |
0.54 |
| chr11_60673508_60673995 | 0.04 |
PRPF19 |
pre-mRNA processing factor 19 |
262 |
0.64 |
| chr19_45393848_45394047 | 0.04 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
121 |
0.58 |
| chr14_102414684_102415292 | 0.04 |
DYNC1H1 |
dynein, cytoplasmic 1, heavy chain 1 |
15877 |
0.19 |
| chr22_21367894_21368045 | 0.04 |
P2RX6 |
purinergic receptor P2X, ligand-gated ion channel, 6 |
1347 |
0.2 |
| chr10_30509279_30509508 | 0.04 |
ENSG00000200887 |
. |
79209 |
0.09 |
| chr22_27068598_27068749 | 0.04 |
CRYBA4 |
crystallin, beta A4 |
50745 |
0.13 |
| chr20_52517333_52517726 | 0.04 |
AC005220.3 |
|
39170 |
0.2 |
| chr5_141404364_141404742 | 0.04 |
GNPDA1 |
glucosamine-6-phosphate deaminase 1 |
11947 |
0.17 |
| chr8_97274172_97274393 | 0.04 |
PTDSS1 |
phosphatidylserine synthase 1 |
141 |
0.8 |
| chr9_127622518_127622669 | 0.04 |
RPL35 |
ribosomal protein L35 |
1601 |
0.24 |
| chr1_10864488_10864639 | 0.04 |
CASZ1 |
castor zinc finger 1 |
7856 |
0.26 |
| chr3_126206096_126206282 | 0.04 |
ZXDC |
ZXD family zinc finger C |
11427 |
0.16 |
| chr12_6798903_6799301 | 0.04 |
ZNF384 |
zinc finger protein 384 |
426 |
0.65 |
| chr1_43637945_43638198 | 0.04 |
WDR65 |
WD repeat domain 65 |
56 |
0.61 |
| chr19_39322550_39322701 | 0.04 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
20 |
0.94 |
| chr21_39228435_39228586 | 0.03 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
57047 |
0.14 |
| chr3_93781580_93781765 | 0.03 |
NSUN3 |
NOP2/Sun domain family, member 3 |
88 |
0.6 |
| chr4_173708323_173708474 | 0.03 |
ENSG00000241652 |
. |
354396 |
0.01 |
| chr1_17914072_17914223 | 0.03 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
764 |
0.76 |
| chr3_28390250_28390542 | 0.03 |
AZI2 |
5-azacytidine induced 2 |
19 |
0.77 |
| chr14_24701887_24702567 | 0.03 |
GMPR2 |
guanosine monophosphate reductase 2 |
73 |
0.8 |
| chr19_58892078_58892419 | 0.03 |
ZNF837 |
zinc finger protein 837 |
141 |
0.87 |
| chr3_88198857_88199168 | 0.03 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
23 |
0.61 |
| chr11_118122819_118123419 | 0.03 |
MPZL3 |
myelin protein zero-like 3 |
54 |
0.96 |
| chr19_51014448_51014741 | 0.03 |
JOSD2 |
Josephin domain containing 2 |
16 |
0.95 |
| chr5_173334437_173334588 | 0.03 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
14431 |
0.25 |
| chr11_134094076_134094379 | 0.03 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
199 |
0.63 |
| chr9_971958_972221 | 0.03 |
DMRT3 |
doublesex and mab-3 related transcription factor 3 |
4875 |
0.26 |
| chr1_32479424_32479717 | 0.03 |
KHDRBS1 |
KH domain containing, RNA binding, signal transduction associated 1 |
27 |
0.97 |
| chr12_51566615_51566911 | 0.03 |
TFCP2 |
transcription factor CP2 |
68 |
0.96 |
| chr19_58570193_58570626 | 0.03 |
ZNF135 |
zinc finger protein 135 |
198 |
0.9 |
| chr12_49961755_49962069 | 0.03 |
MCRS1 |
microspherule protein 1 |
2 |
0.76 |
| chr10_88125065_88125216 | 0.03 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
1095 |
0.59 |
| chr6_144164528_144165302 | 0.03 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
434 |
0.87 |
| chr17_34599013_34599342 | 0.03 |
TBC1D3C |
TBC1 domain family, member 3C |
7145 |
0.15 |
| chr14_21978616_21979591 | 0.03 |
METTL3 |
methyltransferase like 3 |
379 |
0.78 |
| chr4_140216949_140217285 | 0.03 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
160 |
0.93 |
| chr8_122515301_122515452 | 0.03 |
ENSG00000263525 |
. |
39859 |
0.21 |
| chr1_155147010_155147208 | 0.03 |
TRIM46 |
tripartite motif containing 46 |
666 |
0.35 |
| chr10_43932295_43932516 | 0.03 |
ZNF487 |
zinc finger protein 487 |
116 |
0.96 |
| chr19_11616618_11616769 | 0.03 |
ZNF653 |
zinc finger protein 653 |
45 |
0.95 |
| chr3_195946092_195946608 | 0.03 |
SLC51A |
solute carrier family 51, alpha subunit |
1587 |
0.28 |
| chr9_136776234_136776385 | 0.03 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
81094 |
0.08 |
| chr7_108209550_108210056 | 0.03 |
THAP5 |
THAP domain containing 5 |
94 |
0.67 |
| chr10_75504089_75504991 | 0.03 |
SEC24C |
SEC24 family member C |
382 |
0.7 |
| chr1_100231847_100232213 | 0.03 |
FRRS1 |
ferric-chelate reductase 1 |
681 |
0.67 |
| chr15_78592072_78592304 | 0.03 |
WDR61 |
WD repeat domain 61 |
52 |
0.96 |
| chr12_72080266_72080710 | 0.03 |
TMEM19 |
transmembrane protein 19 |
212 |
0.93 |
| chr9_127174465_127174742 | 0.03 |
PSMB7 |
proteasome (prosome, macropain) subunit, beta type, 7 |
3115 |
0.26 |
| chr19_37263819_37264017 | 0.03 |
CTD-2162K18.4 |
Uncharacterized protein |
137 |
0.57 |
| chr3_184033030_184033497 | 0.03 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
67 |
0.94 |
| chr12_49109524_49110644 | 0.03 |
CCNT1 |
cyclin T1 |
597 |
0.6 |
| chr13_110993415_110993566 | 0.03 |
COL4A2 |
collagen, type IV, alpha 2 |
33876 |
0.17 |
| chr5_139724897_139725179 | 0.03 |
HBEGF |
heparin-binding EGF-like growth factor |
1150 |
0.36 |
| chr6_86303657_86304481 | 0.03 |
SNX14 |
sorting nexin 14 |
195 |
0.95 |
| chr2_27028844_27028995 | 0.03 |
CENPA |
centromere protein A |
19983 |
0.16 |
| chr8_6264551_6264771 | 0.03 |
RP11-115C21.2 |
|
2 |
0.91 |
| chr8_55013175_55014052 | 0.03 |
LYPLA1 |
lysophospholipase I |
482 |
0.83 |
| chr12_6649814_6649965 | 0.03 |
RP5-940J5.9 |
|
2353 |
0.11 |
| chr18_8750784_8750935 | 0.03 |
ENSG00000252319 |
. |
30729 |
0.14 |
| chrX_51545867_51547149 | 0.03 |
MAGED1 |
melanoma antigen family D, 1 |
405 |
0.89 |
| chr1_240783288_240783439 | 0.03 |
GREM2 |
gremlin 2, DAN family BMP antagonist |
7914 |
0.2 |
| chr12_58138857_58139538 | 0.03 |
TSPAN31 |
tetraspanin 31 |
32 |
0.92 |
| chr16_31008287_31008527 | 0.03 |
STX1B |
syntaxin 1B |
3869 |
0.1 |
| chr15_43663331_43663561 | 0.03 |
TUBGCP4 |
tubulin, gamma complex associated protein 4 |
133 |
0.62 |
| chr7_41426318_41426600 | 0.03 |
INHBA-AS1 |
INHBA antisense RNA 1 |
307055 |
0.01 |
| chr13_106571784_106572046 | 0.03 |
ENSG00000252550 |
. |
21923 |
0.28 |
| chr16_16043073_16043272 | 0.03 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
262 |
0.93 |
| chr17_37301005_37301156 | 0.03 |
PLXDC1 |
plexin domain containing 1 |
6822 |
0.14 |
| chr2_32502679_32502951 | 0.03 |
YIPF4 |
Yip1 domain family, member 4 |
164 |
0.95 |
| chr9_124048192_124048723 | 0.03 |
GSN |
gelsolin |
413 |
0.59 |
| chr2_43822335_43823890 | 0.03 |
THADA |
thyroid adenoma associated |
1 |
0.98 |
| chr6_27584810_27585278 | 0.03 |
ENSG00000238648 |
. |
14144 |
0.19 |
| chr14_65381486_65381648 | 0.03 |
CHURC1-FNTB |
CHURC1-FNTB readthrough |
356 |
0.32 |
| chr8_94766637_94767113 | 0.03 |
TMEM67 |
transmembrane protein 67 |
197 |
0.9 |
| chr3_28390062_28390213 | 0.02 |
AZI2 |
5-azacytidine induced 2 |
17 |
0.91 |
| chr16_27378896_27379134 | 0.02 |
IL4R |
interleukin 4 receptor |
15019 |
0.19 |
| chr3_45165734_45165944 | 0.02 |
CDCP1 |
CUB domain containing protein 1 |
22075 |
0.2 |
| chr9_132382165_132382732 | 0.02 |
C9orf50 |
chromosome 9 open reading frame 50 |
607 |
0.62 |
| chr7_99006283_99006480 | 0.02 |
PDAP1 |
PDGFA associated protein 1 |
71 |
0.55 |
| chr2_208675899_208676134 | 0.02 |
FZD5 |
frizzled family receptor 5 |
41729 |
0.12 |
| chr11_61892028_61892235 | 0.02 |
INCENP |
inner centromere protein antigens 135/155kDa |
661 |
0.71 |
| chr19_35940283_35940434 | 0.02 |
FFAR2 |
free fatty acid receptor 2 |
259 |
0.87 |
| chr1_20987073_20987907 | 0.02 |
DDOST |
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
391 |
0.83 |
| chr1_148556730_148556881 | 0.02 |
NBPF15 |
neuroblastoma breakpoint family, member 15 |
4038 |
0.3 |
| chr3_42844965_42845933 | 0.02 |
HIGD1A |
HIG1 hypoxia inducible domain family, member 1A |
476 |
0.69 |
| chr3_37901503_37901845 | 0.02 |
ENSG00000235257 |
. |
1537 |
0.34 |
| chr1_220906357_220906642 | 0.02 |
MARC2 |
mitochondrial amidoxime reducing component 2 |
15112 |
0.19 |
| chr8_102120904_102121301 | 0.02 |
ENSG00000202360 |
. |
29085 |
0.21 |
| chr18_3248652_3249060 | 0.02 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
985 |
0.49 |
| chr7_42896707_42897080 | 0.02 |
C7orf25 |
chromosome 7 open reading frame 25 |
54616 |
0.14 |
| chr1_33593144_33593416 | 0.02 |
ADC |
arginine decarboxylase |
45429 |
0.12 |
| chr12_14923072_14924123 | 0.02 |
HIST4H4 |
histone cluster 4, H4 |
468 |
0.69 |
| chr17_34510955_34511411 | 0.02 |
AC131056.3 |
|
342 |
0.79 |
| chr1_33772863_33773014 | 0.02 |
RP11-415J8.3 |
|
29 |
0.96 |
| chrX_146993330_146993481 | 0.02 |
FMR1 |
fragile X mental retardation 1 |
64 |
0.5 |
| chr11_130183263_130184165 | 0.02 |
ZBTB44 |
zinc finger and BTB domain containing 44 |
772 |
0.68 |
| chr5_150163315_150163466 | 0.02 |
AC010441.1 |
|
5523 |
0.16 |
| chr11_57408298_57408449 | 0.02 |
ENSG00000208009 |
. |
298 |
0.8 |
| chr19_40596081_40597156 | 0.02 |
AC005614.3 |
|
43 |
0.64 |
| chr1_169764110_169764261 | 0.02 |
C1orf112 |
chromosome 1 open reading frame 112 |
4 |
0.55 |
| chr15_62682825_62683674 | 0.02 |
RP11-299H22.5 |
|
120370 |
0.05 |
| chr11_809579_809818 | 0.02 |
RPLP2 |
ribosomal protein, large, P2 |
51 |
0.91 |
| chr17_65362498_65362713 | 0.02 |
PSMD12 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
116 |
0.96 |
| chr2_198063557_198063736 | 0.02 |
ANKRD44 |
ankyrin repeat domain 44 |
884 |
0.43 |
| chr5_72599487_72599638 | 0.02 |
TMEM174 |
transmembrane protein 174 |
130540 |
0.04 |
| chr15_57998770_57998921 | 0.02 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
18 |
0.99 |
| chr1_180881031_180881376 | 0.02 |
KIAA1614 |
KIAA1614 |
1122 |
0.51 |
| chr2_163174641_163175537 | 0.02 |
IFIH1 |
interferon induced with helicase C domain 1 |
105 |
0.76 |
| chr15_25684395_25684621 | 0.02 |
UBE3A |
ubiquitin protein ligase E3A |
380 |
0.82 |
| chrX_40976244_40976504 | 0.02 |
USP9X |
ubiquitin specific peptidase 9, X-linked |
31486 |
0.22 |
| chr20_388999_389747 | 0.02 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
386 |
0.82 |
| chr1_216773992_216774229 | 0.02 |
ESRRG |
estrogen-related receptor gamma |
122564 |
0.06 |
| chr12_46385346_46385747 | 0.02 |
SCAF11 |
SR-related CTD-associated factor 11 |
357 |
0.93 |
| chr12_88178270_88178421 | 0.02 |
C12orf50 |
chromosome 12 open reading frame 50 |
243395 |
0.02 |
| chr20_30326525_30327002 | 0.02 |
TPX2 |
TPX2, microtubule-associated |
311 |
0.86 |
| chrX_48754632_48755057 | 0.02 |
TIMM17B |
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
186 |
0.66 |
| chr15_66906109_66906400 | 0.02 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
31726 |
0.13 |
| chr10_116755612_116755763 | 0.02 |
ENSG00000252611 |
. |
26360 |
0.22 |
| chr17_25623130_25623281 | 0.02 |
WSB1 |
WD repeat and SOCS box containing 1 |
1833 |
0.35 |
| chr15_76442977_76443390 | 0.02 |
RP11-593F23.1 |
|
30644 |
0.19 |
| chr3_81545158_81545309 | 0.02 |
ENSG00000222389 |
. |
13394 |
0.32 |
| chr10_131761561_131761767 | 0.02 |
EBF3 |
early B-cell factor 3 |
441 |
0.9 |
| chr19_44331131_44331711 | 0.02 |
ZNF283 |
zinc finger protein 283 |
23 |
0.53 |
| chr5_174151065_174151298 | 0.02 |
MSX2 |
msh homeobox 2 |
355 |
0.92 |
| chr6_50784871_50785022 | 0.02 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
1490 |
0.57 |
| chr8_144416622_144416994 | 0.02 |
TOP1MT |
topoisomerase (DNA) I, mitochondrial |
157 |
0.91 |
| chr12_118745072_118745584 | 0.02 |
TAOK3 |
TAO kinase 3 |
51464 |
0.15 |
| chr2_242576780_242577011 | 0.02 |
THAP4 |
THAP domain containing 4 |
31 |
0.62 |
| chr12_101673896_101674669 | 0.02 |
UTP20 |
UTP20, small subunit (SSU) processome component, homolog (yeast) |
395 |
0.87 |
| chr18_74271433_74271584 | 0.02 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
30634 |
0.18 |
| chr7_5862773_5862988 | 0.02 |
ZNF815P |
zinc finger protein 815, pseudogene |
42 |
0.97 |
| chr4_81189966_81190509 | 0.02 |
FGF5 |
fibroblast growth factor 5 |
2444 |
0.36 |
| chr5_173145147_173145298 | 0.02 |
ENSG00000263401 |
. |
11859 |
0.27 |
| chr21_45209367_45209684 | 0.02 |
RRP1 |
ribosomal RNA processing 1 |
131 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |