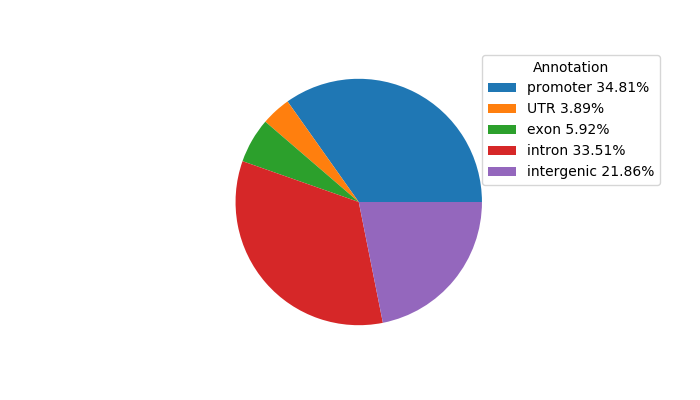
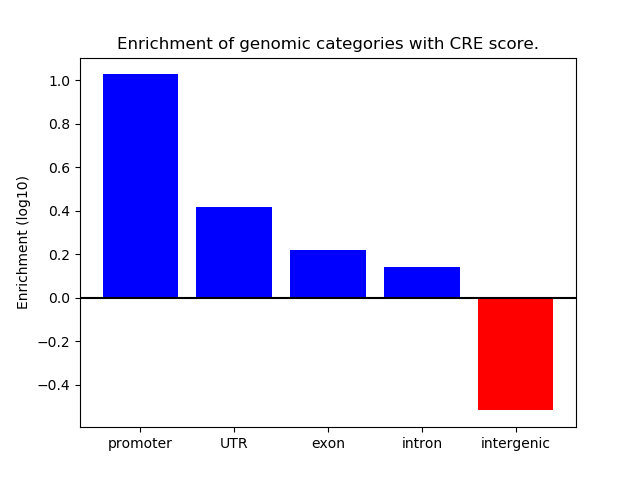
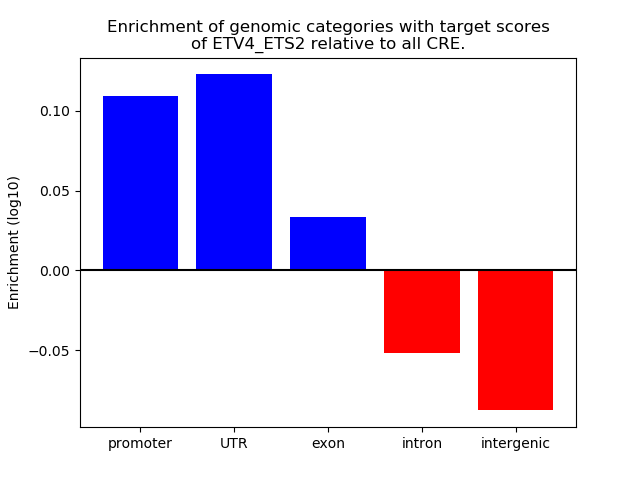
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ETV4_ETS2
Z-value: 0.73
Transcription factors associated with ETV4_ETS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
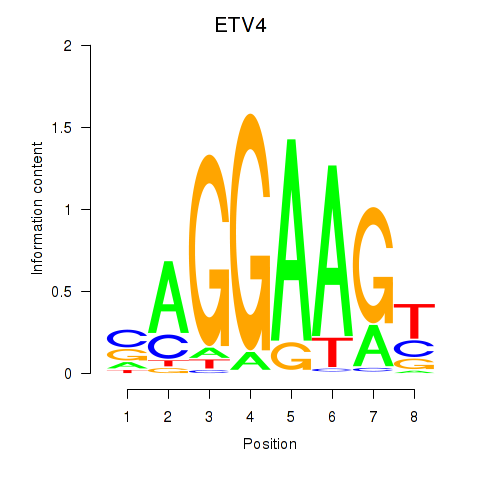
ETV4
|
ENSG00000175832.8 | ETS variant transcription factor 4 |
|
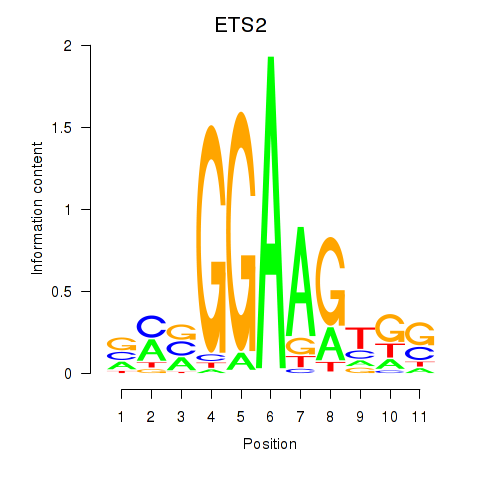
ETS2
|
ENSG00000157557.7 | ETS proto-oncogene 2, transcription factor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr21_40145836_40145987 | ETS2 | 31320 | 0.225744 | -0.63 | 7.0e-02 | Click! |
| chr21_40123884_40124035 | ETS2 | 53272 | 0.157989 | 0.62 | 7.4e-02 | Click! |
| chr21_40211978_40212129 | ETS2 | 30493 | 0.211955 | -0.58 | 1.0e-01 | Click! |
| chr21_40145573_40145724 | ETS2 | 31583 | 0.224902 | -0.57 | 1.1e-01 | Click! |
| chr21_40138917_40139228 | ETS2 | 38159 | 0.203751 | 0.54 | 1.3e-01 | Click! |
| chr17_41623926_41624196 | ETV4 | 261 | 0.873284 | -0.85 | 3.7e-03 | Click! |
| chr17_41622914_41623315 | ETV4 | 33 | 0.946190 | -0.82 | 6.6e-03 | Click! |
| chr17_41669450_41669625 | ETV4 | 12549 | 0.163785 | -0.77 | 1.6e-02 | Click! |
| chr17_41668751_41668949 | ETV4 | 11862 | 0.164855 | -0.77 | 1.6e-02 | Click! |
| chr17_41669802_41669953 | ETV4 | 12889 | 0.163264 | -0.75 | 2.0e-02 | Click! |
Activity of the ETV4_ETS2 motif across conditions
Conditions sorted by the z-value of the ETV4_ETS2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
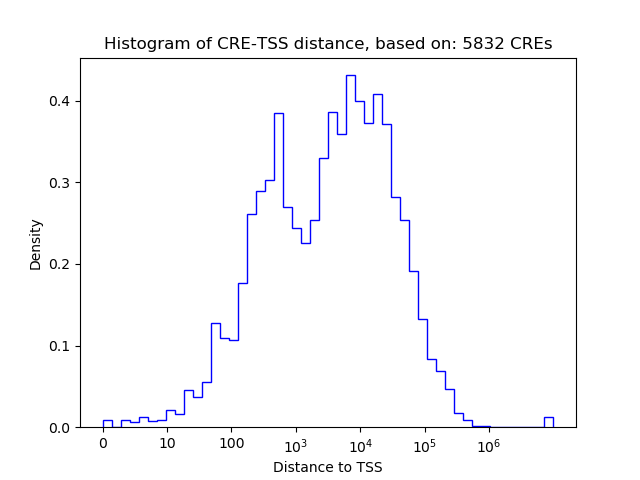
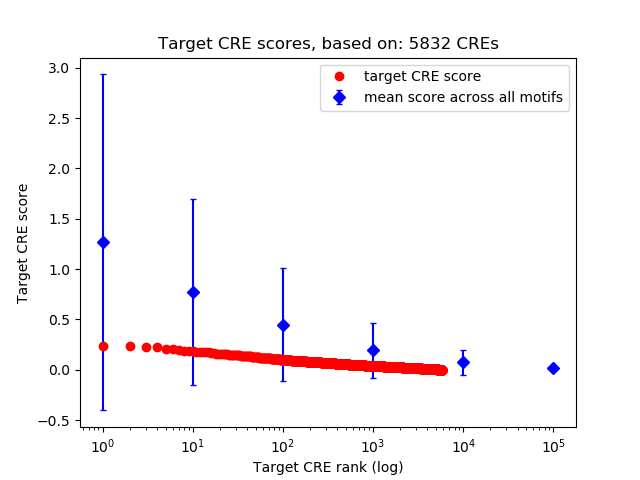
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_63974616_63975431 | 0.24 |
FERMT3 |
fermitin family member 3 |
266 |
0.78 |
| chr12_51714745_51715007 | 0.24 |
BIN2 |
bridging integrator 2 |
3023 |
0.21 |
| chr9_95857151_95857367 | 0.23 |
RP11-274J16.5 |
|
552 |
0.64 |
| chr9_95726568_95726886 | 0.23 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
484 |
0.84 |
| chr11_65407442_65407916 | 0.21 |
SIPA1 |
signal-induced proliferation-associated 1 |
87 |
0.92 |
| chr10_73531555_73531706 | 0.21 |
C10orf54 |
chromosome 10 open reading frame 54 |
1625 |
0.38 |
| chr4_2800484_2801318 | 0.20 |
SH3BP2 |
SH3-domain binding protein 2 |
176 |
0.95 |
| chr22_50319474_50319638 | 0.19 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
7175 |
0.17 |
| chr1_245215310_245215673 | 0.19 |
ENSG00000251754 |
. |
8261 |
0.17 |
| chr17_29814991_29815971 | 0.18 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
355 |
0.83 |
| chr17_7239932_7240900 | 0.18 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
507 |
0.51 |
| chr1_167586551_167586989 | 0.18 |
RCSD1 |
RCSD domain containing 1 |
12560 |
0.19 |
| chr10_75624935_75625135 | 0.18 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
4423 |
0.13 |
| chr14_101587281_101587508 | 0.18 |
ENSG00000206761 |
. |
24638 |
0.04 |
| chr13_46752366_46752713 | 0.18 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
3920 |
0.19 |
| chrX_129225814_129226276 | 0.17 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
18291 |
0.19 |
| chr17_4618141_4618477 | 0.16 |
ARRB2 |
arrestin, beta 2 |
575 |
0.53 |
| chr19_1067222_1068216 | 0.16 |
HMHA1 |
histocompatibility (minor) HA-1 |
222 |
0.85 |
| chr20_62708665_62709256 | 0.16 |
RGS19 |
regulator of G-protein signaling 19 |
1885 |
0.17 |
| chr3_13050979_13051187 | 0.16 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
22547 |
0.24 |
| chr1_79111525_79111735 | 0.16 |
IFI44 |
interferon-induced protein 44 |
3886 |
0.26 |
| chr13_99195211_99195547 | 0.16 |
STK24 |
serine/threonine kinase 24 |
21127 |
0.2 |
| chr19_49839028_49839359 | 0.16 |
CD37 |
CD37 molecule |
472 |
0.63 |
| chr1_160676943_160677601 | 0.15 |
CD48 |
CD48 molecule |
4321 |
0.18 |
| chr3_52279791_52280791 | 0.15 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
66 |
0.94 |
| chr11_128587302_128587453 | 0.15 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
21459 |
0.17 |
| chr17_66344269_66344555 | 0.15 |
ARSG |
arylsulfatase G |
56753 |
0.1 |
| chr7_100484820_100485063 | 0.15 |
SRRT |
serrate RNA effector molecule homolog (Arabidopsis) |
383 |
0.71 |
| chr10_126300385_126300576 | 0.15 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
91714 |
0.07 |
| chr15_102182223_102182374 | 0.15 |
ENSG00000252614 |
. |
6217 |
0.18 |
| chr1_27945148_27945299 | 0.15 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
5350 |
0.15 |
| chr11_60932018_60932169 | 0.14 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
3004 |
0.22 |
| chr19_55084758_55085279 | 0.14 |
LILRA2 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
200 |
0.63 |
| chr10_11212951_11213685 | 0.14 |
RP3-323N1.2 |
|
21 |
0.98 |
| chr11_6251374_6251525 | 0.14 |
FAM160A2 |
family with sequence similarity 160, member A2 |
4397 |
0.13 |
| chr1_206732541_206732911 | 0.14 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
2233 |
0.27 |
| chr12_54866933_54867431 | 0.14 |
GTSF1 |
gametocyte specific factor 1 |
201 |
0.92 |
| chr2_106392629_106392859 | 0.14 |
NCK2 |
NCK adaptor protein 2 |
30556 |
0.23 |
| chr2_175498105_175498391 | 0.14 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
1059 |
0.58 |
| chr6_144980278_144980429 | 0.13 |
UTRN |
utrophin |
627 |
0.85 |
| chr7_150329208_150329551 | 0.13 |
GIMAP6 |
GTPase, IMAP family member 6 |
55 |
0.98 |
| chr7_50727890_50728167 | 0.13 |
GRB10 |
growth factor receptor-bound protein 10 |
33401 |
0.21 |
| chr18_74836287_74836734 | 0.13 |
MBP |
myelin basic protein |
3158 |
0.38 |
| chr19_1895348_1895705 | 0.13 |
SCAMP4 |
secretory carrier membrane protein 4 |
9687 |
0.09 |
| chr17_38478388_38479657 | 0.13 |
RARA |
retinoic acid receptor, alpha |
4485 |
0.13 |
| chr7_63361181_63361806 | 0.13 |
ENSG00000263891 |
. |
47 |
0.98 |
| chr8_125210334_125210485 | 0.13 |
FER1L6-AS2 |
FER1L6 antisense RNA 2 |
26646 |
0.21 |
| chr8_144328075_144328422 | 0.13 |
ZFP41 |
ZFP41 zinc finger protein |
743 |
0.53 |
| chr8_97324979_97325228 | 0.13 |
ENSG00000199732 |
. |
3553 |
0.24 |
| chr10_94590680_94591107 | 0.13 |
EXOC6 |
exocyst complex component 6 |
42 |
0.99 |
| chr16_29674660_29674967 | 0.13 |
QPRT |
quinolinate phosphoribosyltransferase |
213 |
0.51 |
| chr16_17542920_17543071 | 0.13 |
XYLT1 |
xylosyltransferase I |
21743 |
0.29 |
| chr14_101034010_101035263 | 0.12 |
BEGAIN |
brain-enriched guanylate kinase-associated |
192 |
0.93 |
| chr9_117145588_117145751 | 0.12 |
AKNA |
AT-hook transcription factor |
4574 |
0.23 |
| chr10_7526759_7527322 | 0.12 |
ENSG00000207453 |
. |
1731 |
0.45 |
| chr8_145086794_145086945 | 0.12 |
PARP10 |
poly (ADP-ribose) polymerase family, member 10 |
71 |
0.74 |
| chr13_46752006_46752233 | 0.12 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
4340 |
0.19 |
| chr6_111180837_111181007 | 0.12 |
ENSG00000199360 |
. |
3302 |
0.2 |
| chr6_32160504_32160787 | 0.12 |
GPSM3 |
G-protein signaling modulator 3 |
0 |
0.94 |
| chr2_220113848_220114166 | 0.12 |
STK16 |
serine/threonine kinase 16 |
3389 |
0.09 |
| chr16_79123602_79124194 | 0.12 |
RP11-556H2.3 |
|
49 |
0.98 |
| chr16_29756670_29756842 | 0.12 |
AC009133.17 |
|
188 |
0.77 |
| chr21_15914339_15914644 | 0.12 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
4171 |
0.29 |
| chr5_14594534_14594878 | 0.12 |
FAM105A |
family with sequence similarity 105, member A |
12822 |
0.24 |
| chr10_11187446_11187644 | 0.12 |
CELF2 |
CUGBP, Elav-like family member 2 |
19448 |
0.19 |
| chr11_47396682_47396833 | 0.12 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
3185 |
0.13 |
| chr1_203734376_203734572 | 0.12 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
169 |
0.94 |
| chr6_166747166_166747317 | 0.11 |
SFT2D1 |
SFT2 domain containing 1 |
8838 |
0.19 |
| chr20_34078435_34078749 | 0.11 |
RP3-477O4.14 |
|
151 |
0.92 |
| chr1_28213912_28214063 | 0.11 |
THEMIS2 |
thymocyte selection associated family member 2 |
7745 |
0.12 |
| chr20_57465496_57466112 | 0.11 |
GNAS |
GNAS complex locus |
553 |
0.66 |
| chr9_134150778_134151758 | 0.11 |
FAM78A |
family with sequence similarity 78, member A |
666 |
0.69 |
| chr4_26263054_26263205 | 0.11 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
11100 |
0.31 |
| chr3_111851637_111852199 | 0.11 |
GCSAM |
germinal center-associated, signaling and motility |
154 |
0.73 |
| chr4_1287250_1287401 | 0.11 |
MAEA |
macrophage erythroblast attacher |
3642 |
0.18 |
| chr2_175357999_175358525 | 0.11 |
GPR155 |
G protein-coupled receptor 155 |
6440 |
0.21 |
| chr5_150594214_150594453 | 0.11 |
GM2A |
GM2 ganglioside activator |
2622 |
0.27 |
| chr5_544016_544540 | 0.11 |
ENSG00000264233 |
. |
8281 |
0.14 |
| chr19_49859286_49859437 | 0.11 |
AC010524.4 |
|
48 |
0.94 |
| chr1_26616865_26617016 | 0.11 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
10327 |
0.11 |
| chr19_13206703_13207270 | 0.11 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
6695 |
0.11 |
| chr1_198648918_198649184 | 0.11 |
RP11-553K8.5 |
|
12861 |
0.25 |
| chr9_95532616_95532767 | 0.11 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
5597 |
0.21 |
| chr20_47377126_47377429 | 0.11 |
ENSG00000251876 |
. |
21292 |
0.24 |
| chr17_2697553_2697769 | 0.11 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
2071 |
0.29 |
| chr9_126628860_126629011 | 0.11 |
DENND1A |
DENN/MADD domain containing 1A |
63451 |
0.12 |
| chr19_35645615_35646695 | 0.11 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
310 |
0.8 |
| chr19_10715319_10715578 | 0.11 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
2262 |
0.17 |
| chrX_117832511_117832662 | 0.11 |
IL13RA1 |
interleukin 13 receptor, alpha 1 |
28949 |
0.19 |
| chr17_34603649_34603800 | 0.10 |
TBC1D3C |
TBC1 domain family, member 3C |
11692 |
0.14 |
| chr19_42210710_42210868 | 0.10 |
CEACAM5 |
carcinoembryonic antigen-related cell adhesion molecule 5 |
1715 |
0.23 |
| chr1_36042987_36043882 | 0.10 |
RP4-728D4.2 |
|
104 |
0.95 |
| chrX_70328757_70329094 | 0.10 |
IL2RG |
interleukin 2 receptor, gamma |
193 |
0.89 |
| chr11_117841514_117841665 | 0.10 |
IL10RA |
interleukin 10 receptor, alpha |
15474 |
0.17 |
| chr9_95787859_95788010 | 0.10 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
10592 |
0.19 |
| chr6_106611529_106611909 | 0.10 |
RP1-134E15.3 |
|
63704 |
0.11 |
| chr11_1874347_1875032 | 0.10 |
LSP1 |
lymphocyte-specific protein 1 |
489 |
0.65 |
| chr9_117146231_117146573 | 0.10 |
AKNA |
AT-hook transcription factor |
3841 |
0.25 |
| chr19_48747671_48748205 | 0.10 |
CARD8 |
caspase recruitment domain family, member 8 |
3618 |
0.15 |
| chr5_172272383_172272534 | 0.10 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
11044 |
0.19 |
| chr22_27067492_27067685 | 0.10 |
CRYBA4 |
crystallin, beta A4 |
49660 |
0.13 |
| chr5_148620092_148620243 | 0.10 |
ABLIM3 |
actin binding LIM protein family, member 3 |
2744 |
0.22 |
| chr5_176882439_176883305 | 0.10 |
DBN1 |
drebrin 1 |
6518 |
0.1 |
| chr6_41168130_41168571 | 0.10 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
582 |
0.65 |
| chr4_148713676_148713874 | 0.10 |
ENSG00000264274 |
. |
10029 |
0.19 |
| chrX_39949292_39950247 | 0.10 |
BCOR |
BCL6 corepressor |
6887 |
0.33 |
| chr7_119172_119323 | 0.10 |
FAM20C |
family with sequence similarity 20, member C |
73722 |
0.11 |
| chr1_2480724_2480985 | 0.10 |
TNFRSF14 |
tumor necrosis factor receptor superfamily, member 14 |
6224 |
0.1 |
| chr7_73625229_73625469 | 0.10 |
LAT2 |
linker for activation of T cells family, member 2 |
996 |
0.49 |
| chr2_242556723_242557374 | 0.10 |
THAP4 |
THAP domain containing 4 |
132 |
0.94 |
| chr6_14857980_14858173 | 0.10 |
ENSG00000206960 |
. |
211310 |
0.02 |
| chr15_40598587_40598738 | 0.10 |
PLCB2 |
phospholipase C, beta 2 |
1364 |
0.23 |
| chr20_37434091_37434759 | 0.10 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
58 |
0.98 |
| chr10_79362617_79362768 | 0.10 |
ENSG00000199592 |
. |
15885 |
0.23 |
| chr19_50400390_50400724 | 0.09 |
IL4I1 |
interleukin 4 induced 1 |
345 |
0.67 |
| chr5_95159606_95160004 | 0.09 |
GLRX |
glutaredoxin (thioltransferase) |
1096 |
0.44 |
| chr20_23063904_23064055 | 0.09 |
CD93 |
CD93 molecule |
2998 |
0.23 |
| chr2_112913175_112913326 | 0.09 |
FBLN7 |
fibulin 7 |
4075 |
0.29 |
| chr7_6065805_6065956 | 0.09 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
303 |
0.79 |
| chr16_30915229_30915507 | 0.09 |
CTF1 |
cardiotrophin 1 |
7409 |
0.08 |
| chr3_13455515_13455928 | 0.09 |
NUP210 |
nucleoporin 210kDa |
6088 |
0.26 |
| chr1_11800239_11800390 | 0.09 |
AGTRAP |
angiotensin II receptor-associated protein |
4082 |
0.15 |
| chr17_34617167_34617318 | 0.09 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
8477 |
0.15 |
| chr3_53201143_53201294 | 0.09 |
PRKCD |
protein kinase C, delta |
2073 |
0.29 |
| chr1_153363293_153363489 | 0.09 |
S100A8 |
S100 calcium binding protein A8 |
61 |
0.95 |
| chr13_99930587_99930738 | 0.09 |
GPR18 |
G protein-coupled receptor 18 |
16664 |
0.18 |
| chr22_37637445_37637596 | 0.09 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
2768 |
0.2 |
| chr2_69062344_69062572 | 0.09 |
AC097495.2 |
|
322 |
0.9 |
| chr11_122587919_122588091 | 0.09 |
ENSG00000239079 |
. |
9014 |
0.25 |
| chr13_25855828_25855979 | 0.09 |
MTMR6 |
myotubularin related protein 6 |
5801 |
0.2 |
| chr7_36325944_36326095 | 0.09 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
10734 |
0.18 |
| chr5_131810697_131810848 | 0.09 |
AC116366.6 |
|
2037 |
0.23 |
| chr2_8612888_8613039 | 0.09 |
AC011747.7 |
|
202933 |
0.03 |
| chr2_232573344_232574118 | 0.09 |
PTMA |
prothymosin, alpha |
485 |
0.75 |
| chr8_59506492_59506643 | 0.09 |
SDCBP |
syndecan binding protein (syntenin) |
28992 |
0.19 |
| chr17_72459969_72460120 | 0.09 |
CD300A |
CD300a molecule |
2511 |
0.22 |
| chr14_51280333_51280548 | 0.09 |
RP11-286O18.1 |
|
8158 |
0.15 |
| chr19_51631094_51631245 | 0.09 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
882 |
0.38 |
| chr5_175123540_175123739 | 0.09 |
ENSG00000200648 |
. |
7262 |
0.21 |
| chr11_67176589_67176876 | 0.09 |
TBC1D10C |
TBC1 domain family, member 10C |
5072 |
0.07 |
| chr13_33607800_33607951 | 0.09 |
ENSG00000221677 |
. |
7614 |
0.24 |
| chr12_15104428_15104579 | 0.09 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
418 |
0.81 |
| chr15_99192776_99194432 | 0.09 |
IGF1R |
insulin-like growth factor 1 receptor |
1331 |
0.47 |
| chr10_134365004_134365155 | 0.09 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
13436 |
0.21 |
| chr6_36666192_36666416 | 0.09 |
RAB44 |
RAB44, member RAS oncogene family |
16952 |
0.13 |
| chr2_169961988_169962139 | 0.09 |
AC007556.3 |
|
4810 |
0.25 |
| chr9_102587867_102588202 | 0.09 |
NR4A3 |
nuclear receptor subfamily 4, group A, member 3 |
975 |
0.66 |
| chr19_10982465_10983821 | 0.09 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
764 |
0.53 |
| chr21_36900755_36901002 | 0.09 |
ENSG00000211590 |
. |
192135 |
0.03 |
| chr8_125208186_125208337 | 0.09 |
FER1L6-AS2 |
FER1L6 antisense RNA 2 |
24498 |
0.22 |
| chr1_203254106_203254257 | 0.09 |
BTG2 |
BTG family, member 2 |
20483 |
0.15 |
| chr9_117149358_117150269 | 0.09 |
AKNA |
AT-hook transcription factor |
430 |
0.85 |
| chr2_149303870_149304072 | 0.09 |
MBD5 |
methyl-CpG binding domain protein 5 |
77677 |
0.11 |
| chr15_69961298_69961449 | 0.09 |
ENSG00000238870 |
. |
61788 |
0.13 |
| chr5_131793340_131793491 | 0.09 |
ENSG00000202533 |
. |
10424 |
0.13 |
| chr15_40659564_40659826 | 0.09 |
RP11-64K12.4 |
|
2146 |
0.13 |
| chr17_75437754_75438017 | 0.09 |
ENSG00000200651 |
. |
306 |
0.87 |
| chr9_120477806_120477957 | 0.09 |
ENSG00000201444 |
. |
7501 |
0.21 |
| chr4_169427075_169427226 | 0.09 |
PALLD |
palladin, cytoskeletal associated protein |
5569 |
0.22 |
| chr6_143177934_143178085 | 0.09 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
19825 |
0.26 |
| chr20_19055744_19055895 | 0.09 |
ENSG00000264669 |
. |
50610 |
0.16 |
| chr6_35279100_35279566 | 0.09 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1818 |
0.34 |
| chr7_158505123_158505274 | 0.09 |
NCAPG2 |
non-SMC condensin II complex, subunit G2 |
7678 |
0.23 |
| chr3_71493443_71493594 | 0.09 |
ENSG00000221264 |
. |
97722 |
0.08 |
| chr19_30171358_30171509 | 0.08 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
13652 |
0.21 |
| chr1_204482334_204482501 | 0.08 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
3094 |
0.23 |
| chr17_27220072_27220391 | 0.08 |
RP11-20B24.7 |
|
744 |
0.4 |
| chr7_106505707_106506635 | 0.08 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
247 |
0.95 |
| chr21_48042285_48042436 | 0.08 |
PRMT2 |
protein arginine methyltransferase 2 |
12719 |
0.2 |
| chr7_137620698_137620944 | 0.08 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
2185 |
0.34 |
| chr9_101881133_101881552 | 0.08 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
8715 |
0.21 |
| chrX_17848143_17848294 | 0.08 |
RAI2 |
retinoic acid induced 2 |
7632 |
0.3 |
| chr7_138757613_138757764 | 0.08 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
6325 |
0.22 |
| chr6_13636625_13636776 | 0.08 |
AL441883.1 |
Uncharacterized protein |
15574 |
0.14 |
| chr5_156696070_156696221 | 0.08 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
217 |
0.91 |
| chr8_30514582_30515646 | 0.08 |
GTF2E2 |
general transcription factor IIE, polypeptide 2, beta 34kDa |
21 |
0.98 |
| chr7_2684869_2685165 | 0.08 |
TTYH3 |
tweety family member 3 |
149 |
0.96 |
| chr10_112619332_112619483 | 0.08 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
12158 |
0.14 |
| chr7_26415747_26416790 | 0.08 |
AC004540.4 |
|
53 |
0.98 |
| chr8_102150052_102150203 | 0.08 |
ENSG00000202360 |
. |
60 |
0.98 |
| chr10_70804041_70804192 | 0.08 |
SRGN |
serglycin |
43758 |
0.12 |
| chr7_148844568_148845756 | 0.08 |
ZNF398 |
zinc finger protein 398 |
57 |
0.97 |
| chrX_38676446_38676597 | 0.08 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
13385 |
0.23 |
| chr11_64510621_64511071 | 0.08 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
415 |
0.76 |
| chr9_139117966_139118117 | 0.08 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
1318 |
0.43 |
| chr5_175665565_175666307 | 0.08 |
SIMC1 |
SUMO-interacting motifs containing 1 |
533 |
0.81 |
| chr2_70171863_70172014 | 0.08 |
ENSG00000239072 |
. |
10839 |
0.12 |
| chr11_75479321_75480435 | 0.08 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
21 |
0.69 |
| chr9_117690313_117690616 | 0.08 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
2233 |
0.42 |
| chr10_27148230_27149879 | 0.08 |
ABI1 |
abl-interactor 1 |
738 |
0.67 |
| chr11_64546452_64546665 | 0.08 |
SF1 |
splicing factor 1 |
300 |
0.83 |
| chr1_8285162_8285313 | 0.08 |
ENSG00000200975 |
. |
18580 |
0.19 |
| chr1_6524907_6525205 | 0.08 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
634 |
0.58 |
| chr11_64107746_64107914 | 0.08 |
CCDC88B |
coiled-coil domain containing 88B |
135 |
0.9 |
| chr18_9095215_9095557 | 0.08 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
7242 |
0.18 |
| chr11_133826464_133827261 | 0.08 |
IGSF9B |
immunoglobulin superfamily, member 9B |
18 |
0.58 |
| chr5_40616837_40616988 | 0.08 |
ENSG00000199552 |
. |
38153 |
0.18 |
| chr1_2509263_2509520 | 0.08 |
RP3-395M20.9 |
|
3608 |
0.12 |
| chr17_81147720_81147871 | 0.08 |
METRNL |
meteorin, glial cell differentiation regulator-like |
95801 |
0.08 |
| chr8_28918241_28918392 | 0.08 |
CTD-2647L4.4 |
|
4864 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0002864 | regulation of acute inflammatory response to antigenic stimulus(GO:0002864) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0002517 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0002643 | regulation of tolerance induction(GO:0002643) positive regulation of tolerance induction(GO:0002645) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |