Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
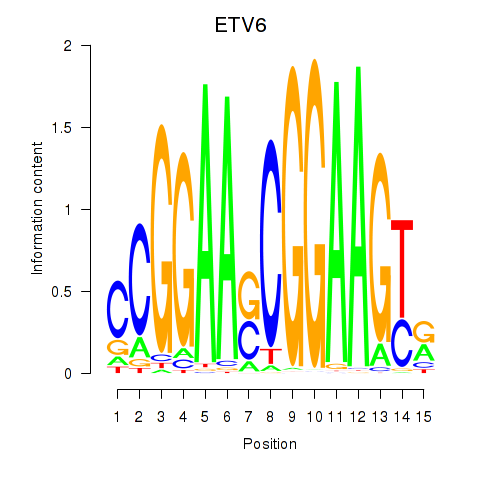
Results for ETV6
Z-value: 0.68
Transcription factors associated with ETV6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV6
|
ENSG00000139083.6 | ETS variant transcription factor 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_11922803_11922954 | ETV6 | 17443 | 0.275566 | 0.76 | 1.7e-02 | Click! |
| chr12_11808723_11808874 | ETV6 | 6010 | 0.290192 | 0.73 | 2.5e-02 | Click! |
| chr12_11883028_11883179 | ETV6 | 22332 | 0.256967 | 0.72 | 2.8e-02 | Click! |
| chr12_11877911_11878062 | ETV6 | 27449 | 0.240572 | 0.70 | 3.6e-02 | Click! |
| chr12_11883469_11883620 | ETV6 | 21891 | 0.258351 | 0.69 | 4.0e-02 | Click! |
Activity of the ETV6 motif across conditions
Conditions sorted by the z-value of the ETV6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
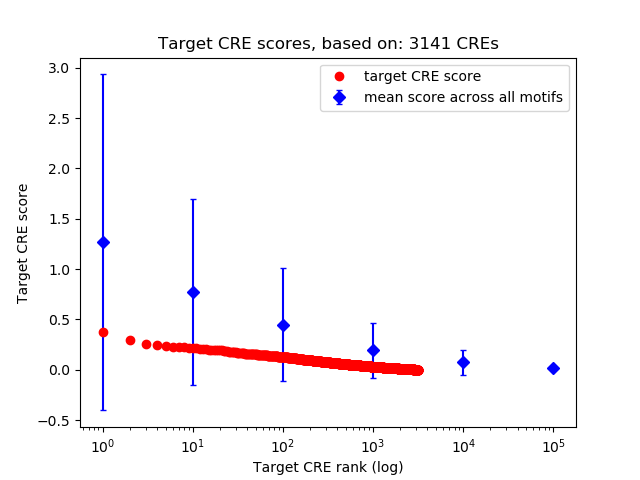
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_53693277_53694636 | 0.37 |
C12orf10 |
chromosome 12 open reading frame 10 |
79 |
0.58 |
| chr4_185735910_185736201 | 0.30 |
RP11-701P16.2 |
Uncharacterized protein |
1282 |
0.41 |
| chr20_23064192_23064486 | 0.26 |
CD93 |
CD93 molecule |
2638 |
0.25 |
| chr9_126774768_126775305 | 0.25 |
LHX2 |
LIM homeobox 2 |
400 |
0.79 |
| chr6_37017654_37018643 | 0.23 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
5541 |
0.22 |
| chr13_32621993_32622144 | 0.23 |
FRY |
furry homolog (Drosophila) |
12933 |
0.22 |
| chr6_33610158_33610417 | 0.23 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
21126 |
0.13 |
| chr2_63274875_63275396 | 0.22 |
AC009501.4 |
|
289 |
0.91 |
| chr12_122075303_122075715 | 0.22 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
10788 |
0.17 |
| chr14_24787929_24788080 | 0.22 |
LTB4R |
leukotriene B4 receptor |
4098 |
0.08 |
| chr17_65430423_65430574 | 0.22 |
ENSG00000244610 |
. |
22908 |
0.12 |
| chr16_17156854_17157148 | 0.21 |
CTD-2576D5.4 |
|
71360 |
0.14 |
| chr15_102182223_102182374 | 0.21 |
ENSG00000252614 |
. |
6217 |
0.18 |
| chr19_457823_457974 | 0.20 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
3098 |
0.14 |
| chr8_22571062_22571213 | 0.20 |
EGR3 |
early growth response 3 |
20322 |
0.13 |
| chr19_15740026_15740177 | 0.20 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
11606 |
0.18 |
| chr22_39350720_39350871 | 0.19 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
2039 |
0.24 |
| chr15_91459401_91459658 | 0.19 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
3411 |
0.11 |
| chr10_99094156_99094718 | 0.19 |
FRAT2 |
frequently rearranged in advanced T-cell lymphomas 2 |
21 |
0.6 |
| chr10_121166225_121166376 | 0.19 |
GRK5 |
G protein-coupled receptor kinase 5 |
28351 |
0.17 |
| chr1_43397747_43398281 | 0.19 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
1218 |
0.47 |
| chr16_89396583_89396806 | 0.19 |
ANKRD11 |
ankyrin repeat domain 11 |
1300 |
0.35 |
| chr9_117134390_117135168 | 0.19 |
AKNA |
AT-hook transcription factor |
4465 |
0.23 |
| chr15_90602890_90603041 | 0.19 |
ZNF710 |
zinc finger protein 710 |
8281 |
0.15 |
| chr11_118057234_118057385 | 0.18 |
SCN2B |
sodium channel, voltage-gated, type II, beta subunit |
9921 |
0.14 |
| chr19_46525700_46525851 | 0.18 |
PGLYRP1 |
peptidoglycan recognition protein 1 |
548 |
0.66 |
| chr19_46094450_46094601 | 0.18 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
6425 |
0.11 |
| chr17_38473365_38473678 | 0.18 |
RARA |
retinoic acid receptor, alpha |
1012 |
0.41 |
| chrX_18743813_18743964 | 0.18 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
18111 |
0.2 |
| chr3_36934586_36934737 | 0.17 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
34287 |
0.18 |
| chr7_97755628_97755779 | 0.17 |
LMTK2 |
lemur tyrosine kinase 2 |
19506 |
0.2 |
| chr1_38465132_38465538 | 0.17 |
FHL3 |
four and a half LIM domains 3 |
5842 |
0.13 |
| chr5_10445338_10445751 | 0.17 |
ROPN1L |
rhophilin associated tail protein 1-like |
3556 |
0.18 |
| chr19_49838375_49839003 | 0.17 |
CD37 |
CD37 molecule |
5 |
0.95 |
| chr1_17519453_17519604 | 0.16 |
PADI1 |
peptidyl arginine deiminase, type I |
12093 |
0.17 |
| chr9_139927070_139927409 | 0.16 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
223 |
0.78 |
| chr10_72163983_72165283 | 0.16 |
EIF4EBP2 |
eukaryotic translation initiation factor 4E binding protein 2 |
498 |
0.77 |
| chr5_50695310_50695461 | 0.16 |
ISL1 |
ISL LIM homeobox 1 |
15879 |
0.22 |
| chr19_50143768_50144112 | 0.16 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
482 |
0.54 |
| chr4_31024444_31024595 | 0.16 |
RP11-619J20.1 |
|
228927 |
0.02 |
| chr11_64624166_64624317 | 0.16 |
CDC42BPG |
CDC42 binding protein kinase gamma (DMPK-like) |
12200 |
0.1 |
| chr2_119130108_119130259 | 0.16 |
INSIG2 |
insulin induced gene 2 |
284133 |
0.01 |
| chr22_19879311_19879851 | 0.16 |
GNB1L |
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
37119 |
0.1 |
| chr19_8645617_8645768 | 0.16 |
AC130469.2 |
|
352 |
0.8 |
| chr19_19483852_19484190 | 0.16 |
GATAD2A |
GATA zinc finger domain containing 2A |
12614 |
0.13 |
| chr2_8627682_8628105 | 0.16 |
AC011747.7 |
|
188003 |
0.03 |
| chr21_34442470_34442979 | 0.16 |
OLIG1 |
oligodendrocyte transcription factor 1 |
274 |
0.77 |
| chr5_137090212_137090730 | 0.15 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
432 |
0.85 |
| chr6_28227027_28227486 | 0.15 |
NKAPL |
NFKB activating protein-like |
158 |
0.61 |
| chr20_42166672_42166823 | 0.15 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
9400 |
0.15 |
| chr9_127266363_127266826 | 0.15 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
2920 |
0.28 |
| chr22_42832160_42832442 | 0.15 |
NFAM1 |
NFAT activating protein with ITAM motif 1 |
3900 |
0.23 |
| chr16_10479736_10481071 | 0.15 |
ATF7IP2 |
activating transcription factor 7 interacting protein 2 |
485 |
0.83 |
| chr19_48833533_48834100 | 0.15 |
EMP3 |
epithelial membrane protein 3 |
4951 |
0.12 |
| chr16_28505762_28506015 | 0.15 |
APOBR |
apolipoprotein B receptor |
82 |
0.93 |
| chr4_40623487_40623692 | 0.15 |
RBM47 |
RNA binding motif protein 47 |
8292 |
0.27 |
| chr9_132646855_132647649 | 0.15 |
FNBP1 |
formin binding protein 1 |
34337 |
0.12 |
| chr21_36077269_36077677 | 0.15 |
CLIC6 |
chloride intracellular channel 6 |
35785 |
0.17 |
| chr14_103588344_103588755 | 0.15 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
1249 |
0.43 |
| chr1_87244034_87244185 | 0.15 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
73531 |
0.09 |
| chr15_101708121_101708357 | 0.15 |
RP11-505E24.2 |
|
81968 |
0.08 |
| chr17_40993833_40993984 | 0.15 |
AOC2 |
amine oxidase, copper containing 2 (retina-specific) |
2709 |
0.12 |
| chr3_133293335_133294350 | 0.15 |
CDV3 |
CDV3 homolog (mouse) |
350 |
0.89 |
| chr16_4909761_4909912 | 0.15 |
UBN1 |
ubinuclein 1 |
7620 |
0.14 |
| chr17_73584494_73585142 | 0.15 |
MYO15B |
myosin XVB pseudogene |
2040 |
0.21 |
| chr19_55084758_55085279 | 0.14 |
LILRA2 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
200 |
0.63 |
| chr17_45772156_45772908 | 0.14 |
TBKBP1 |
TBK1 binding protein 1 |
98 |
0.95 |
| chr15_30213515_30213794 | 0.14 |
TJP1 |
tight junction protein 1 |
47414 |
0.15 |
| chr11_62767141_62767292 | 0.14 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
14761 |
0.14 |
| chr21_36217976_36218127 | 0.14 |
RUNX1 |
runt-related transcription factor 1 |
41429 |
0.2 |
| chr1_205717555_205718736 | 0.14 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
1259 |
0.38 |
| chr5_112539683_112539834 | 0.14 |
MCC |
mutated in colorectal cancers |
30616 |
0.23 |
| chr14_23588365_23588516 | 0.14 |
CEBPE |
CCAAT/enhancer binding protein (C/EBP), epsilon |
385 |
0.75 |
| chr20_6947669_6947820 | 0.14 |
ENSG00000251833 |
. |
106083 |
0.08 |
| chr16_9207976_9208127 | 0.14 |
RP11-473I1.10 |
|
6953 |
0.16 |
| chr1_32123629_32123780 | 0.14 |
RP11-73M7.6 |
|
845 |
0.49 |
| chr11_8190407_8190734 | 0.14 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
2 |
0.98 |
| chr1_12244133_12244323 | 0.14 |
ENSG00000263676 |
. |
7542 |
0.16 |
| chr19_4374031_4374264 | 0.14 |
SH3GL1 |
SH3-domain GRB2-like 1 |
6142 |
0.08 |
| chr6_129944528_129944679 | 0.14 |
ARHGAP18 |
Rho GTPase activating protein 18 |
86767 |
0.09 |
| chr1_22351811_22352015 | 0.14 |
ENSG00000201273 |
. |
14435 |
0.09 |
| chr14_70111468_70111619 | 0.14 |
KIAA0247 |
KIAA0247 |
33230 |
0.18 |
| chr16_84766426_84766761 | 0.13 |
USP10 |
ubiquitin specific peptidase 10 |
32947 |
0.16 |
| chr1_223306949_223307134 | 0.13 |
TLR5 |
toll-like receptor 5 |
1057 |
0.67 |
| chr16_69373556_69374479 | 0.13 |
NIP7 |
NIP7, nucleolar pre-rRNA processing protein |
157 |
0.76 |
| chr8_121457410_121457992 | 0.13 |
MTBP |
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
35 |
0.54 |
| chr21_34905254_34905405 | 0.13 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
9091 |
0.13 |
| chr2_7571269_7571803 | 0.13 |
ENSG00000221255 |
. |
145436 |
0.05 |
| chr20_56048279_56048430 | 0.13 |
CTCFL |
CCCTC-binding factor (zinc finger protein)-like |
50959 |
0.11 |
| chr6_130341588_130342312 | 0.13 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
248 |
0.96 |
| chr16_85786290_85786441 | 0.13 |
C16orf74 |
chromosome 16 open reading frame 74 |
1630 |
0.24 |
| chr3_47418814_47418965 | 0.13 |
RP11-708J19.1 |
|
3600 |
0.2 |
| chr2_28817013_28817303 | 0.13 |
PLB1 |
phospholipase B1 |
7628 |
0.24 |
| chr19_56166514_56167932 | 0.13 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
792 |
0.34 |
| chr11_19990149_19990300 | 0.13 |
NAV2-AS3 |
NAV2 antisense RNA 3 |
12659 |
0.24 |
| chr1_46859625_46860587 | 0.13 |
FAAH |
fatty acid amide hydrolase |
169 |
0.95 |
| chr3_15332172_15332523 | 0.13 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
14795 |
0.14 |
| chr8_61764729_61764880 | 0.13 |
RP11-33I11.2 |
|
42639 |
0.2 |
| chr19_1754756_1754961 | 0.13 |
ONECUT3 |
one cut homeobox 3 |
2486 |
0.2 |
| chr19_12902481_12903520 | 0.13 |
JUNB |
jun B proto-oncogene |
690 |
0.39 |
| chr12_4380012_4380209 | 0.13 |
CCND2 |
cyclin D2 |
2828 |
0.22 |
| chr3_167812746_167813857 | 0.13 |
GOLIM4 |
golgi integral membrane protein 4 |
169 |
0.97 |
| chr4_7060277_7060428 | 0.13 |
TADA2B |
transcriptional adaptor 2B |
5272 |
0.13 |
| chr19_17439012_17439348 | 0.13 |
ANO8 |
anoctamin 8 |
6458 |
0.08 |
| chr12_7055489_7056183 | 0.12 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
63 |
0.9 |
| chr21_16429371_16429522 | 0.12 |
NRIP1 |
nuclear receptor interacting protein 1 |
7680 |
0.28 |
| chr7_6066625_6066776 | 0.12 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
517 |
0.61 |
| chr11_130030115_130030717 | 0.12 |
ST14 |
suppression of tumorigenicity 14 (colon carcinoma) |
959 |
0.66 |
| chr3_49943448_49943849 | 0.12 |
CTD-2330K9.3 |
Uncharacterized protein |
2228 |
0.16 |
| chr5_139926895_139927116 | 0.12 |
EIF4EBP3 |
eukaryotic translation initiation factor 4E binding protein 3 |
246 |
0.82 |
| chr2_38151836_38153080 | 0.12 |
RMDN2 |
regulator of microtubule dynamics 2 |
4 |
0.99 |
| chr16_70487949_70488475 | 0.12 |
FUK |
fucokinase |
112 |
0.94 |
| chr11_57566897_57567048 | 0.12 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
7941 |
0.18 |
| chr4_56261758_56262564 | 0.12 |
TMEM165 |
transmembrane protein 165 |
31 |
0.65 |
| chr6_225379_225671 | 0.12 |
DUSP22 |
dual specificity phosphatase 22 |
66105 |
0.15 |
| chr1_161008749_161009060 | 0.12 |
TSTD1 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
124 |
0.91 |
| chr2_242674070_242675162 | 0.12 |
AC114730.8 |
|
297 |
0.65 |
| chr16_23132061_23132212 | 0.12 |
USP31 |
ubiquitin specific peptidase 31 |
28455 |
0.2 |
| chr19_38756015_38756259 | 0.12 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
159 |
0.9 |
| chr17_57408824_57410302 | 0.12 |
YPEL2 |
yippee-like 2 (Drosophila) |
500 |
0.78 |
| chr12_94519902_94520172 | 0.12 |
PLXNC1 |
plexin C1 |
22462 |
0.21 |
| chr16_11707122_11707273 | 0.12 |
LITAF |
lipopolysaccharide-induced TNF factor |
14841 |
0.18 |
| chr5_171840008_171840159 | 0.12 |
ENSG00000216127 |
. |
26675 |
0.19 |
| chr17_29149849_29150048 | 0.12 |
CRLF3 |
cytokine receptor-like factor 3 |
1741 |
0.25 |
| chr9_137258561_137258730 | 0.12 |
ENSG00000263897 |
. |
12612 |
0.24 |
| chr7_37487305_37488000 | 0.12 |
ELMO1 |
engulfment and cell motility 1 |
901 |
0.61 |
| chr22_35772700_35773071 | 0.12 |
HMOX1 |
heme oxygenase (decycling) 1 |
3469 |
0.23 |
| chr1_156630677_156631393 | 0.12 |
RP11-284F21.7 |
|
181 |
0.89 |
| chr2_54643569_54643720 | 0.12 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
39778 |
0.18 |
| chr11_128554784_128555228 | 0.11 |
RP11-744N12.3 |
|
1317 |
0.34 |
| chr17_54910275_54910729 | 0.11 |
C17orf67 |
chromosome 17 open reading frame 67 |
754 |
0.45 |
| chr10_22540455_22541288 | 0.11 |
EBLN1 |
endogenous Bornavirus-like nucleoprotein 1 |
41921 |
0.14 |
| chr13_37005266_37006411 | 0.11 |
CCNA1 |
cyclin A1 |
129 |
0.97 |
| chr12_10205099_10205456 | 0.11 |
CLEC9A |
C-type lectin domain family 9, member A |
22001 |
0.1 |
| chr14_107081796_107081947 | 0.11 |
IGHV4-59 |
immunoglobulin heavy variable 4-59 |
1854 |
0.07 |
| chr17_49337942_49338261 | 0.11 |
UTP18 |
UTP18 small subunit (SSU) processome component homolog (yeast) |
212 |
0.82 |
| chr1_154980554_154980872 | 0.11 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
5418 |
0.08 |
| chr6_130848326_130848477 | 0.11 |
ENSG00000202438 |
. |
46856 |
0.17 |
| chr19_51522714_51522865 | 0.11 |
KLK10 |
kallikrein-related peptidase 10 |
231 |
0.81 |
| chr22_33636979_33637130 | 0.11 |
RP1-302D9.3 |
|
77005 |
0.1 |
| chr9_109649535_109649686 | 0.11 |
ZNF462 |
zinc finger protein 462 |
24163 |
0.19 |
| chr17_36715319_36715684 | 0.11 |
SRCIN1 |
SRC kinase signaling inhibitor 1 |
4159 |
0.19 |
| chr7_74277653_74277804 | 0.11 |
GTF2IRD2 |
GTF2I repeat domain containing 2 |
9881 |
0.18 |
| chr6_12483851_12484268 | 0.11 |
ENSG00000223321 |
. |
77046 |
0.12 |
| chr14_100842881_100843420 | 0.11 |
WARS |
tryptophanyl-tRNA synthetase |
8 |
0.78 |
| chr19_51631094_51631245 | 0.11 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
882 |
0.38 |
| chr13_24845503_24846606 | 0.11 |
SPATA13 |
spermatogenesis associated 13 |
773 |
0.61 |
| chr9_95878549_95878700 | 0.11 |
NINJ1 |
ninjurin 1 |
17946 |
0.14 |
| chr19_911654_912010 | 0.11 |
R3HDM4 |
R3H domain containing 4 |
1399 |
0.21 |
| chr11_1328574_1328800 | 0.11 |
TOLLIP |
toll interacting protein |
2162 |
0.23 |
| chr19_10380429_10381098 | 0.11 |
ICAM1 |
intercellular adhesion molecule 1 |
748 |
0.39 |
| chr6_130702998_130703205 | 0.11 |
TMEM200A |
transmembrane protein 200A |
16222 |
0.2 |
| chr11_129695482_129695633 | 0.11 |
TMEM45B |
transmembrane protein 45B |
9843 |
0.27 |
| chr12_13251108_13251431 | 0.11 |
GSG1 |
germ cell associated 1 |
2529 |
0.3 |
| chr1_24069439_24069920 | 0.11 |
TCEB3 |
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
34 |
0.96 |
| chr1_230262662_230262881 | 0.11 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
59753 |
0.13 |
| chr2_149303386_149303537 | 0.11 |
MBD5 |
methyl-CpG binding domain protein 5 |
77167 |
0.11 |
| chr7_150329208_150329551 | 0.11 |
GIMAP6 |
GTPase, IMAP family member 6 |
55 |
0.98 |
| chr16_75058643_75058911 | 0.11 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
22582 |
0.18 |
| chr4_140481827_140481978 | 0.11 |
RP11-342I1.2 |
|
3949 |
0.22 |
| chr20_61559769_61560616 | 0.10 |
DIDO1 |
death inducer-obliterator 1 |
2337 |
0.23 |
| chr6_108349048_108349199 | 0.10 |
ENSG00000238490 |
. |
32899 |
0.14 |
| chr1_46073171_46073322 | 0.10 |
NASP |
nuclear autoantigenic sperm protein (histone-binding) |
5595 |
0.17 |
| chr11_60934798_60934949 | 0.10 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
5784 |
0.17 |
| chr9_132406808_132406959 | 0.10 |
ASB6 |
ankyrin repeat and SOCS box containing 6 |
2439 |
0.18 |
| chr16_53128120_53128271 | 0.10 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
4877 |
0.27 |
| chr14_35809151_35809404 | 0.10 |
PSMA6 |
proteasome (prosome, macropain) subunit, alpha type, 6 |
29238 |
0.18 |
| chr14_66602242_66602393 | 0.10 |
ENSG00000200860 |
. |
42036 |
0.2 |
| chr2_219496883_219497034 | 0.10 |
RP11-548H3.1 |
|
1779 |
0.19 |
| chr2_65039482_65039633 | 0.10 |
ENSG00000239891 |
. |
4955 |
0.21 |
| chr1_17508352_17508670 | 0.10 |
PADI1 |
peptidyl arginine deiminase, type I |
23110 |
0.15 |
| chr15_99105917_99106068 | 0.10 |
FAM169B |
family with sequence similarity 169, member B |
48381 |
0.16 |
| chr7_48136018_48136265 | 0.10 |
UPP1 |
uridine phosphorylase 1 |
7168 |
0.24 |
| chr4_15004104_15004915 | 0.10 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
211 |
0.97 |
| chr20_45946936_45948261 | 0.10 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
352 |
0.82 |
| chr7_157130148_157131060 | 0.10 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
367 |
0.9 |
| chr17_77778016_77778167 | 0.10 |
CBX8 |
chromobox homolog 8 |
2759 |
0.18 |
| chr11_46698234_46698478 | 0.10 |
ATG13 |
autophagy related 13 |
12806 |
0.13 |
| chr19_8559121_8559517 | 0.10 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
8176 |
0.12 |
| chr10_91055374_91055594 | 0.10 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
6228 |
0.15 |
| chr12_22840824_22841065 | 0.10 |
ETNK1 |
ethanolamine kinase 1 |
62653 |
0.13 |
| chr7_66146644_66146983 | 0.10 |
RP4-756H11.3 |
|
27240 |
0.16 |
| chr18_47003055_47003217 | 0.10 |
RP11-110H1.4 |
|
9383 |
0.08 |
| chr22_40882106_40882345 | 0.10 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
22787 |
0.15 |
| chr8_61851903_61852054 | 0.10 |
CLVS1 |
clavesin 1 |
117739 |
0.06 |
| chr17_46048416_46049087 | 0.10 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
234 |
0.85 |
| chr15_91427302_91427634 | 0.10 |
FES |
feline sarcoma oncogene |
174 |
0.9 |
| chr13_32607825_32607976 | 0.10 |
FRY-AS1 |
FRY antisense RNA 1 |
2124 |
0.32 |
| chr11_554769_555059 | 0.10 |
RP11-496I9.1 |
|
2681 |
0.11 |
| chr12_50925398_50925549 | 0.10 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
26583 |
0.19 |
| chrX_20283128_20284271 | 0.10 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
1061 |
0.63 |
| chr2_101911542_101911693 | 0.10 |
RNF149 |
ring finger protein 149 |
13541 |
0.16 |
| chr19_48827788_48827939 | 0.10 |
EMP3 |
epithelial membrane protein 3 |
719 |
0.53 |
| chr1_161197360_161197771 | 0.10 |
ENSG00000263548 |
. |
589 |
0.45 |
| chr15_85278174_85278822 | 0.09 |
ZNF592 |
zinc finger protein 592 |
13368 |
0.12 |
| chr3_148995184_148995347 | 0.09 |
RP11-206M11.7 |
|
7304 |
0.21 |
| chr1_209979523_209979948 | 0.09 |
IRF6 |
interferon regulatory factor 6 |
270 |
0.89 |
| chr6_127664293_127665233 | 0.09 |
ECHDC1 |
enoyl CoA hydratase domain containing 1 |
9 |
0.98 |
| chr1_55266980_55267187 | 0.09 |
TTC22 |
tetratricopeptide repeat domain 22 |
143 |
0.94 |
| chr11_118122819_118123419 | 0.09 |
MPZL3 |
myelin protein zero-like 3 |
54 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0052257 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0035987 | endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0072124 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:1902808 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.0 | GO:0022009 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0002639 | positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.0 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |