Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
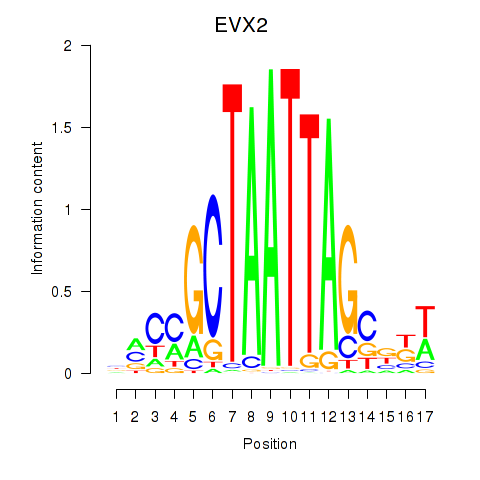
Results for EVX2
Z-value: 0.60
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | even-skipped homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_176932607_176932906 | EVX2 | 15885 | 0.098539 | -0.81 | 7.9e-03 | Click! |
| chr2_176931780_176932252 | EVX2 | 16625 | 0.098417 | -0.56 | 1.2e-01 | Click! |
| chr2_176952907_176953136 | EVX2 | 4380 | 0.100133 | -0.55 | 1.3e-01 | Click! |
| chr2_176943993_176944195 | EVX2 | 4547 | 0.110379 | -0.54 | 1.3e-01 | Click! |
| chr2_176932269_176932561 | EVX2 | 16226 | 0.098484 | -0.47 | 2.0e-01 | Click! |
Activity of the EVX2 motif across conditions
Conditions sorted by the z-value of the EVX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
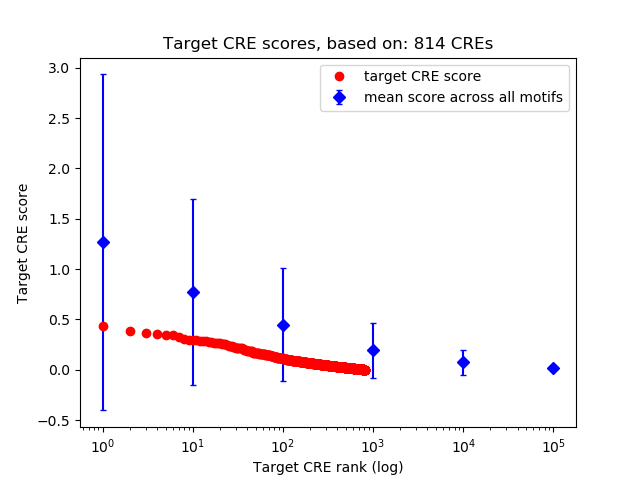
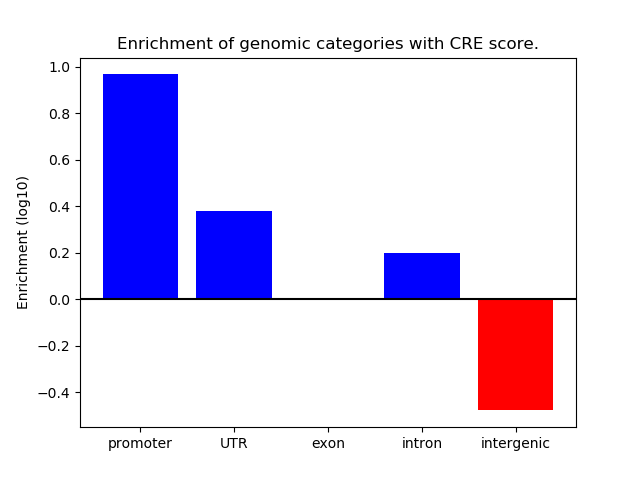
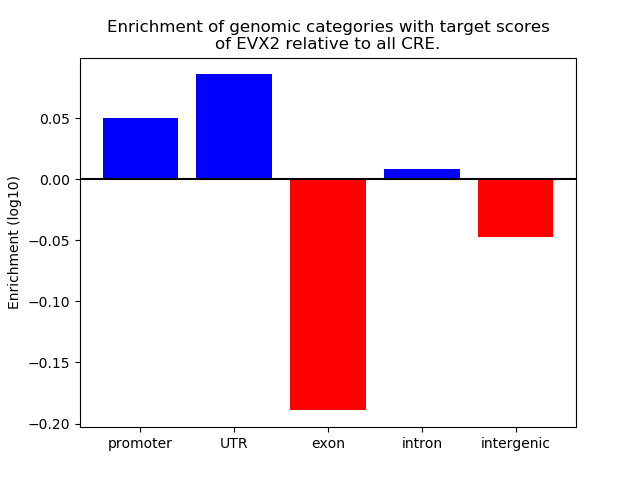
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_74964898_74965186 | 0.44 |
ENSG00000267568 |
. |
137 |
0.97 |
| chr1_234544831_234544982 | 0.39 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
35459 |
0.12 |
| chr2_193992778_193992929 | 0.36 |
TMEFF2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
932418 |
0.0 |
| chr11_128589269_128589524 | 0.36 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chr1_236284065_236284436 | 0.35 |
GPR137B |
G protein-coupled receptor 137B |
21582 |
0.19 |
| chr12_131796680_131796831 | 0.34 |
ENSG00000212251 |
. |
12459 |
0.23 |
| chr4_144312492_144312643 | 0.32 |
GAB1 |
GRB2-associated binding protein 1 |
96 |
0.98 |
| chr7_41921705_41921953 | 0.31 |
AC005027.3 |
|
176896 |
0.03 |
| chr3_32994754_32995260 | 0.29 |
CCR4 |
chemokine (C-C motif) receptor 4 |
1941 |
0.43 |
| chr1_226919798_226919959 | 0.29 |
ITPKB |
inositol-trisphosphate 3-kinase B |
5281 |
0.26 |
| chr16_50776896_50777610 | 0.29 |
RP11-327F22.1 |
|
527 |
0.39 |
| chr9_95790679_95791025 | 0.29 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
13510 |
0.18 |
| chrX_78403286_78403508 | 0.29 |
GPR174 |
G protein-coupled receptor 174 |
23072 |
0.28 |
| chr6_25041553_25042305 | 0.29 |
RP3-425P12.5 |
|
138 |
0.72 |
| chr12_47600587_47601314 | 0.28 |
PCED1B |
PC-esterase domain containing 1B |
9102 |
0.22 |
| chr15_52023462_52024365 | 0.28 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
6405 |
0.15 |
| chr7_115668295_115668446 | 0.27 |
TFEC |
transcription factor EC |
2425 |
0.43 |
| chr4_143321296_143321548 | 0.27 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
30990 |
0.26 |
| chr5_126094279_126094440 | 0.27 |
ENSG00000252185 |
. |
3250 |
0.29 |
| chr16_79652209_79652360 | 0.26 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
17673 |
0.26 |
| chr16_17164825_17164976 | 0.26 |
CTD-2576D5.4 |
|
63461 |
0.16 |
| chr12_64798382_64799443 | 0.25 |
XPOT |
exportin, tRNA |
86 |
0.97 |
| chr1_206646546_206646697 | 0.25 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
2783 |
0.21 |
| chr14_99703180_99703331 | 0.25 |
AL109767.1 |
|
26030 |
0.19 |
| chr20_47440971_47441122 | 0.24 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
3374 |
0.34 |
| chr1_28975271_28975442 | 0.24 |
ENSG00000270103 |
. |
244 |
0.88 |
| chr4_105887659_105887892 | 0.24 |
ENSG00000251906 |
. |
8044 |
0.31 |
| chr13_21050204_21050392 | 0.23 |
ENSG00000263978 |
. |
42313 |
0.15 |
| chr15_70384069_70384220 | 0.23 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
2980 |
0.31 |
| chr12_27150397_27150548 | 0.22 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
2271 |
0.24 |
| chr13_99957828_99957979 | 0.22 |
GPR183 |
G protein-coupled receptor 183 |
1756 |
0.38 |
| chr1_186291714_186291865 | 0.22 |
ENSG00000202025 |
. |
10729 |
0.17 |
| chr22_40322756_40322951 | 0.22 |
GRAP2 |
GRB2-related adaptor protein 2 |
212 |
0.93 |
| chr10_112632655_112633305 | 0.21 |
PDCD4-AS1 |
PDCD4 antisense RNA 1 |
989 |
0.38 |
| chr18_9404580_9404814 | 0.21 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
69849 |
0.08 |
| chr7_37011253_37011404 | 0.20 |
ELMO1 |
engulfment and cell motility 1 |
13337 |
0.2 |
| chr1_150294067_150294674 | 0.20 |
PRPF3 |
pre-mRNA processing factor 3 |
368 |
0.78 |
| chr4_74571443_74571594 | 0.19 |
IL8 |
interleukin 8 |
34705 |
0.18 |
| chr20_47441337_47441845 | 0.19 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
2829 |
0.36 |
| chr2_100388320_100388562 | 0.19 |
AFF3 |
AF4/FMR2 family, member 3 |
193609 |
0.03 |
| chr19_16770752_16770951 | 0.19 |
CTC-429P9.4 |
Small integral membrane protein 7; Uncharacterized protein |
64 |
0.53 |
| chr6_36008402_36008553 | 0.19 |
MAPK14 |
mitogen-activated protein kinase 14 |
11611 |
0.19 |
| chr8_131954085_131954236 | 0.19 |
RP11-737F9.1 |
|
6505 |
0.26 |
| chr10_70850849_70851349 | 0.18 |
SRGN |
serglycin |
3225 |
0.25 |
| chr1_90101335_90101580 | 0.18 |
LRRC8C |
leucine rich repeat containing 8 family, member C |
2826 |
0.22 |
| chr14_93552039_93552233 | 0.18 |
ITPK1-AS1 |
ITPK1 antisense RNA 1 |
18339 |
0.18 |
| chr10_54463336_54463751 | 0.17 |
RP11-556E13.1 |
|
51626 |
0.16 |
| chr8_110619590_110619741 | 0.17 |
SYBU |
syntabulin (syntaxin-interacting) |
639 |
0.72 |
| chr4_139635357_139635508 | 0.17 |
ENSG00000238971 |
. |
97488 |
0.08 |
| chrX_38728231_38728382 | 0.17 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
65170 |
0.13 |
| chr1_225610206_225610357 | 0.17 |
LBR |
lamin B receptor |
5318 |
0.2 |
| chr5_154465051_154465202 | 0.16 |
ENSG00000221131 |
. |
60766 |
0.13 |
| chr4_154412529_154412680 | 0.16 |
KIAA0922 |
KIAA0922 |
25103 |
0.22 |
| chr7_83932213_83932364 | 0.16 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
107527 |
0.08 |
| chr6_143263290_143263441 | 0.16 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
2973 |
0.34 |
| chr6_131948545_131949340 | 0.16 |
MED23 |
mediator complex subunit 23 |
258 |
0.77 |
| chr9_72745702_72745853 | 0.16 |
MAMDC2-AS1 |
MAMDC2 antisense RNA 1 |
17001 |
0.22 |
| chr3_16883621_16883873 | 0.16 |
PLCL2 |
phospholipase C-like 2 |
42705 |
0.19 |
| chr4_1005974_1006310 | 0.16 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
97 |
0.95 |
| chr5_75600050_75600201 | 0.16 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr1_112283056_112283209 | 0.15 |
FAM212B-AS1 |
FAM212B antisense RNA 1 |
657 |
0.55 |
| chr16_53133173_53134292 | 0.15 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
660 |
0.77 |
| chr20_56286148_56286403 | 0.15 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
266 |
0.95 |
| chr7_17039488_17039788 | 0.15 |
AGR3 |
anterior gradient 3 |
118027 |
0.06 |
| chr22_50908966_50909262 | 0.15 |
SBF1 |
SET binding factor 1 |
4257 |
0.1 |
| chr14_39736543_39737240 | 0.15 |
CTAGE5 |
CTAGE family, member 5 |
273 |
0.72 |
| chrX_23969833_23969984 | 0.15 |
CXorf58 |
chromosome X open reading frame 58 |
41408 |
0.14 |
| chr1_90290694_90290845 | 0.15 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
3289 |
0.28 |
| chr18_46455022_46455234 | 0.15 |
SMAD7 |
SMAD family member 7 |
19747 |
0.23 |
| chr4_84031409_84031639 | 0.15 |
PLAC8 |
placenta-specific 8 |
528 |
0.83 |
| chr20_34330600_34331178 | 0.14 |
RBM39 |
RNA binding motif protein 39 |
655 |
0.59 |
| chr17_27252886_27253120 | 0.14 |
RP11-20B24.5 |
|
290 |
0.78 |
| chr1_10516682_10516833 | 0.14 |
RP5-1113E3.3 |
|
1855 |
0.2 |
| chr5_110560941_110561277 | 0.14 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1325 |
0.51 |
| chr3_150258880_150259179 | 0.14 |
SERP1 |
stress-associated endoplasmic reticulum protein 1 |
5257 |
0.22 |
| chr13_83262553_83263100 | 0.14 |
ENSG00000222791 |
. |
609476 |
0.0 |
| chr14_22993528_22993679 | 0.13 |
TRAJ15 |
T cell receptor alpha joining 15 |
4977 |
0.12 |
| chr15_43985556_43986365 | 0.13 |
CKMT1A |
creatine kinase, mitochondrial 1A |
109 |
0.93 |
| chr15_75080868_75081404 | 0.13 |
ENSG00000264386 |
. |
38 |
0.76 |
| chr11_23886778_23886929 | 0.13 |
ENSG00000252519 |
. |
15529 |
0.31 |
| chr4_41884194_41884831 | 0.13 |
TMEM33 |
transmembrane protein 33 |
52625 |
0.14 |
| chr14_70102783_70102934 | 0.13 |
KIAA0247 |
KIAA0247 |
24545 |
0.21 |
| chr14_65192350_65192501 | 0.13 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
1926 |
0.38 |
| chr15_43885730_43886468 | 0.13 |
CKMT1B |
creatine kinase, mitochondrial 1B |
42 |
0.95 |
| chr13_40229560_40229742 | 0.12 |
COG6 |
component of oligomeric golgi complex 6 |
113 |
0.96 |
| chr16_50623254_50623506 | 0.12 |
RP11-401P9.6 |
|
24215 |
0.14 |
| chr19_16482178_16482409 | 0.12 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
9529 |
0.15 |
| chr17_9313239_9313390 | 0.12 |
AC087501.1 |
|
42200 |
0.15 |
| chr7_6114490_6114641 | 0.12 |
ENSG00000264605 |
. |
8171 |
0.11 |
| chr12_94805998_94806149 | 0.12 |
CCDC41 |
coiled-coil domain containing 41 |
47649 |
0.14 |
| chr1_21349011_21349508 | 0.12 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
28136 |
0.19 |
| chr7_30388343_30388494 | 0.12 |
ENSG00000207771 |
. |
59008 |
0.11 |
| chr4_166361643_166362016 | 0.12 |
CPE |
carboxypeptidase E |
22451 |
0.23 |
| chr1_30417491_30417642 | 0.12 |
ENSG00000222787 |
. |
59817 |
0.17 |
| chr6_79788386_79788977 | 0.12 |
PHIP |
pleckstrin homology domain interacting protein |
728 |
0.79 |
| chr14_22747289_22747440 | 0.12 |
ENSG00000238634 |
. |
136477 |
0.04 |
| chr21_15917916_15918619 | 0.12 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr2_112368119_112368270 | 0.11 |
ENSG00000266063 |
. |
160517 |
0.04 |
| chr16_6001852_6002003 | 0.11 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
67168 |
0.13 |
| chr15_100881986_100882218 | 0.11 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
108 |
0.98 |
| chr17_64297613_64298278 | 0.11 |
PRKCA |
protein kinase C, alpha |
999 |
0.57 |
| chr4_146101952_146102435 | 0.11 |
OTUD4 |
OTU domain containing 4 |
880 |
0.65 |
| chr4_26887523_26887909 | 0.11 |
STIM2 |
stromal interaction molecule 2 |
24635 |
0.21 |
| chr2_54798401_54798574 | 0.11 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
12956 |
0.19 |
| chr6_130391698_130391967 | 0.11 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
50130 |
0.15 |
| chr5_108208713_108208864 | 0.11 |
ENSG00000207404 |
. |
18350 |
0.23 |
| chr1_150549248_150549415 | 0.11 |
MCL1 |
myeloid cell leukemia sequence 1 (BCL2-related) |
2675 |
0.12 |
| chr17_10601145_10601947 | 0.11 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
635 |
0.41 |
| chrX_78200973_78201449 | 0.10 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
293 |
0.95 |
| chr10_45389082_45389233 | 0.10 |
TMEM72 |
transmembrane protein 72 |
17491 |
0.16 |
| chr5_134370433_134370584 | 0.10 |
PITX1 |
paired-like homeodomain 1 |
5 |
0.97 |
| chrX_131614340_131615550 | 0.10 |
MBNL3 |
muscleblind-like splicing regulator 3 |
8098 |
0.3 |
| chr1_25887536_25887687 | 0.10 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
17540 |
0.19 |
| chr14_49374188_49374339 | 0.10 |
ENSG00000251824 |
. |
36454 |
0.24 |
| chr1_200987607_200987758 | 0.10 |
KIF21B |
kinesin family member 21B |
4854 |
0.21 |
| chr14_99723235_99723386 | 0.10 |
AL109767.1 |
|
5975 |
0.23 |
| chr1_99843003_99843154 | 0.10 |
LPPR4 |
Lipid phosphate phosphatase-related protein type 4 |
78658 |
0.12 |
| chr8_70622919_70623070 | 0.10 |
RP11-102F4.2 |
|
2631 |
0.31 |
| chr2_235359483_235359634 | 0.10 |
ARL4C |
ADP-ribosylation factor-like 4C |
45686 |
0.21 |
| chr12_1613703_1614089 | 0.09 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
25161 |
0.21 |
| chr1_1710161_1710908 | 0.09 |
NADK |
NAD kinase |
305 |
0.86 |
| chr11_16759613_16759958 | 0.09 |
C11orf58 |
chromosome 11 open reading frame 58 |
163 |
0.97 |
| chr2_38822967_38823118 | 0.09 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
4126 |
0.22 |
| chrX_78620829_78620980 | 0.09 |
ITM2A |
integral membrane protein 2A |
1952 |
0.51 |
| chr6_134499074_134500065 | 0.09 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
526 |
0.8 |
| chr12_54891959_54892179 | 0.09 |
NCKAP1L |
NCK-associated protein 1-like |
499 |
0.73 |
| chr7_105912156_105912307 | 0.09 |
NAMPT |
nicotinamide phosphoribosyltransferase |
13136 |
0.21 |
| chr10_33425320_33425471 | 0.09 |
ENSG00000263576 |
. |
37831 |
0.16 |
| chr19_13049368_13049753 | 0.09 |
CALR |
calreticulin |
101 |
0.89 |
| chr8_87357008_87357171 | 0.09 |
WWP1 |
WW domain containing E3 ubiquitin protein ligase 1 |
2122 |
0.42 |
| chr17_75456326_75456913 | 0.09 |
SEPT9 |
septin 9 |
4171 |
0.18 |
| chr15_31690130_31690281 | 0.09 |
KLF13 |
Kruppel-like factor 13 |
31848 |
0.24 |
| chr1_108505298_108505449 | 0.09 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
1692 |
0.35 |
| chr22_21357082_21357447 | 0.09 |
THAP7-AS1 |
THAP7 antisense RNA 1 |
767 |
0.29 |
| chr17_16724813_16724964 | 0.09 |
AC022596.6 |
|
918 |
0.46 |
| chr6_26312991_26313348 | 0.09 |
HIST1H4H |
histone cluster 1, H4h |
27407 |
0.07 |
| chr2_162095067_162095304 | 0.09 |
TANK |
TRAF family member-associated NFKB activator |
7488 |
0.21 |
| chr10_70230757_70231440 | 0.09 |
DNA2 |
DNA replication helicase/nuclease 2 |
545 |
0.71 |
| chr6_30180418_30180721 | 0.09 |
TRIM26 |
tripartite motif containing 26 |
574 |
0.68 |
| chr13_48975408_48975569 | 0.09 |
LPAR6 |
lysophosphatidic acid receptor 6 |
25555 |
0.23 |
| chr2_103506378_103506656 | 0.09 |
TMEM182 |
transmembrane protein 182 |
128045 |
0.06 |
| chr1_121159975_121160126 | 0.09 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
224113 |
0.02 |
| chr13_46627012_46627449 | 0.09 |
CPB2-AS1 |
CPB2 antisense RNA 1 |
193 |
0.67 |
| chr9_80524074_80524374 | 0.09 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chr1_204548801_204548952 | 0.09 |
ENSG00000200408 |
. |
17227 |
0.14 |
| chr11_117151480_117151734 | 0.09 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
14594 |
0.13 |
| chr3_18472261_18472412 | 0.08 |
SATB1 |
SATB homeobox 1 |
4409 |
0.24 |
| chr17_78965248_78965600 | 0.08 |
CHMP6 |
charged multivesicular body protein 6 |
26 |
0.96 |
| chr7_84685147_84685298 | 0.08 |
SEMA3D |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
66029 |
0.15 |
| chr1_200332191_200332460 | 0.08 |
ZNF281 |
zinc finger protein 281 |
46795 |
0.19 |
| chr7_36243067_36243218 | 0.08 |
AC007327.5 |
|
27574 |
0.18 |
| chr1_158786969_158787120 | 0.08 |
MNDA |
myeloid cell nuclear differentiation antigen |
14063 |
0.17 |
| chr10_124269771_124269922 | 0.08 |
HTRA1 |
HtrA serine peptidase 1 |
3639 |
0.26 |
| chr10_105211471_105211777 | 0.08 |
CALHM2 |
calcium homeostasis modulator 2 |
451 |
0.57 |
| chr12_866633_866784 | 0.08 |
WNK1 |
WNK lysine deficient protein kinase 1 |
3976 |
0.25 |
| chr10_90752530_90752985 | 0.08 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
1610 |
0.28 |
| chr1_100820231_100820573 | 0.08 |
CDC14A |
cell division cycle 14A |
1897 |
0.33 |
| chr2_216877074_216877561 | 0.08 |
MREG |
melanoregulin |
1029 |
0.61 |
| chr11_114129890_114130041 | 0.08 |
NNMT |
nicotinamide N-methyltransferase |
1412 |
0.51 |
| chr11_88068800_88069370 | 0.08 |
CTSC |
cathepsin C |
1816 |
0.49 |
| chr2_99552897_99553419 | 0.08 |
KIAA1211L |
KIAA1211-like |
436 |
0.88 |
| chr10_7453140_7453326 | 0.08 |
SFMBT2 |
Scm-like with four mbt domains 2 |
215 |
0.96 |
| chr6_41855450_41855601 | 0.08 |
USP49 |
ubiquitin specific peptidase 49 |
458 |
0.75 |
| chr5_173293780_173293931 | 0.08 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
21428 |
0.25 |
| chr6_130030490_130031674 | 0.08 |
ARHGAP18 |
Rho GTPase activating protein 18 |
288 |
0.94 |
| chr10_63753889_63754171 | 0.08 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
54940 |
0.15 |
| chr15_34875013_34876331 | 0.08 |
GOLGA8B |
golgin A8 family, member B |
99 |
0.96 |
| chr14_91091040_91091191 | 0.08 |
TTC7B |
tetratricopeptide repeat domain 7B |
6748 |
0.2 |
| chr8_38237822_38238929 | 0.08 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
36 |
0.96 |
| chr10_77308241_77308392 | 0.08 |
ENSG00000207583 |
. |
3900 |
0.31 |
| chr10_46075820_46076565 | 0.08 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
13747 |
0.25 |
| chr2_198572065_198572426 | 0.08 |
MARS2 |
methionyl-tRNA synthetase 2, mitochondrial |
2158 |
0.3 |
| chr2_80472785_80472936 | 0.08 |
ENSG00000221179 |
. |
1112 |
0.61 |
| chr11_128582862_128583013 | 0.08 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
17019 |
0.17 |
| chr1_160614457_160614608 | 0.08 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
2279 |
0.26 |
| chr6_90868813_90868964 | 0.07 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
137573 |
0.04 |
| chr10_173487_173638 | 0.07 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
6843 |
0.24 |
| chr7_6654683_6655004 | 0.07 |
ZNF853 |
zinc finger protein 853 |
405 |
0.77 |
| chr3_124774787_124775424 | 0.07 |
HEG1 |
heart development protein with EGF-like domains 1 |
303 |
0.88 |
| chr14_88471433_88472132 | 0.07 |
GPR65 |
G protein-coupled receptor 65 |
314 |
0.88 |
| chr13_91999115_91999698 | 0.07 |
ENSG00000215417 |
. |
3453 |
0.35 |
| chr2_136876136_136876323 | 0.07 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
494 |
0.87 |
| chr17_25622078_25622646 | 0.07 |
WSB1 |
WD repeat and SOCS box containing 1 |
990 |
0.48 |
| chr6_30923060_30923530 | 0.07 |
HCG21 |
HLA complex group 21 (non-protein coding) |
656 |
0.5 |
| chr19_55656842_55656993 | 0.07 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
1393 |
0.22 |
| chr1_111420872_111421052 | 0.07 |
CD53 |
CD53 molecule |
5186 |
0.22 |
| chr18_11852846_11853378 | 0.07 |
RP11-78A19.3 |
|
362 |
0.77 |
| chr5_148931227_148931707 | 0.07 |
ARHGEF37 |
Rho guanine nucleotide exchange factor (GEF) 37 |
43 |
0.88 |
| chr19_6481304_6482171 | 0.07 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr17_39742070_39742221 | 0.07 |
KRT14 |
keratin 14 |
1028 |
0.36 |
| chr14_32672268_32672419 | 0.07 |
ENSG00000202337 |
. |
132 |
0.94 |
| chr8_97273295_97273764 | 0.07 |
MTERFD1 |
MTERF domain containing 1 |
287 |
0.6 |
| chr19_12888779_12888973 | 0.07 |
HOOK2 |
hook microtubule-tethering protein 2 |
350 |
0.68 |
| chr7_5594421_5594572 | 0.07 |
CTB-161C1.1 |
|
1866 |
0.25 |
| chr10_75686047_75686198 | 0.07 |
C10orf55 |
chromosome 10 open reading frame 55 |
3587 |
0.18 |
| chr16_88937052_88937336 | 0.07 |
PABPN1L |
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
4126 |
0.12 |
| chr7_139476140_139477008 | 0.07 |
HIPK2 |
homeodomain interacting protein kinase 2 |
943 |
0.52 |
| chrX_35788537_35788688 | 0.07 |
MAGEB16 |
melanoma antigen family B, 16 |
27847 |
0.25 |
| chr15_50557147_50557298 | 0.07 |
HDC |
histidine decarboxylase |
641 |
0.66 |
| chr6_154568354_154568864 | 0.07 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |