Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
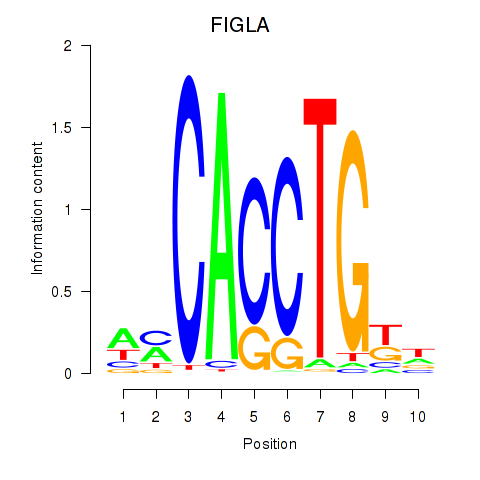
Results for FIGLA
Z-value: 0.20
Transcription factors associated with FIGLA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FIGLA
|
ENSG00000183733.6 | folliculogenesis specific bHLH transcription factor |
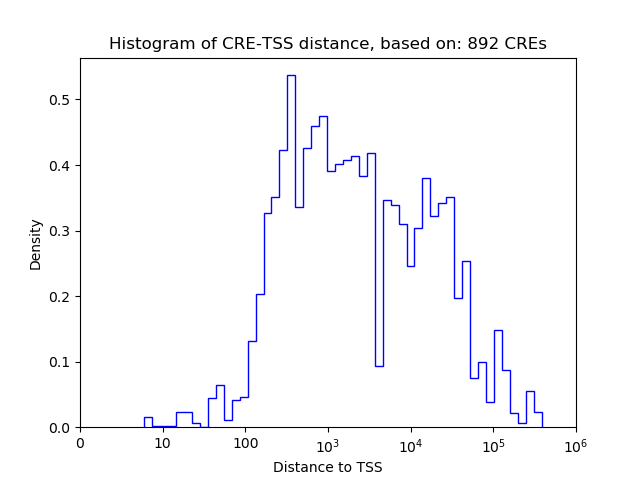
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_71016684_71016881 | FIGLA | 993 | 0.490381 | -0.71 | 3.2e-02 | Click! |
| chr2_71017218_71017686 | FIGLA | 323 | 0.865016 | -0.22 | 5.7e-01 | Click! |
| chr2_71016964_71017169 | FIGLA | 709 | 0.627364 | 0.02 | 9.6e-01 | Click! |
Activity of the FIGLA motif across conditions
Conditions sorted by the z-value of the FIGLA motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
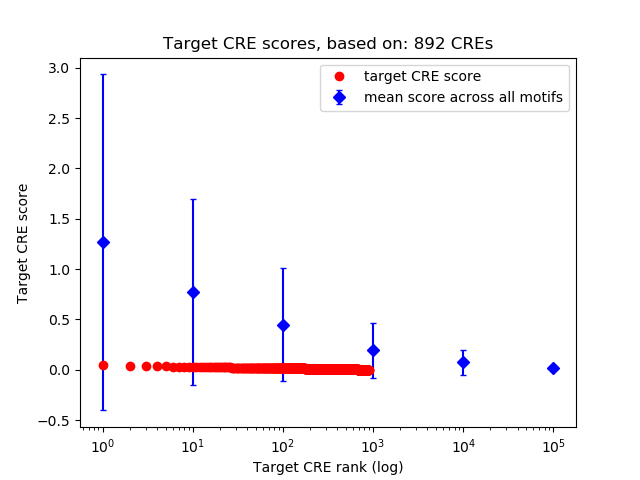
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_86126252_86126683 | 0.05 |
RP11-815J21.2 |
|
3058 |
0.23 |
| chr17_78748885_78749036 | 0.04 |
RP11-28G8.1 |
|
30472 |
0.2 |
| chr9_132802183_132802428 | 0.04 |
FNBP1 |
formin binding protein 1 |
3136 |
0.26 |
| chr2_178217813_178218079 | 0.04 |
ENSG00000238295 |
. |
7257 |
0.15 |
| chr13_24827167_24827318 | 0.04 |
SPATA13-AS1 |
SPATA13 antisense RNA 1 |
1335 |
0.33 |
| chr18_55448427_55448643 | 0.03 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
21792 |
0.18 |
| chr17_8867884_8868296 | 0.03 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
934 |
0.65 |
| chr4_40203939_40204397 | 0.03 |
RHOH |
ras homolog family member H |
2204 |
0.34 |
| chr5_133458466_133458617 | 0.03 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
768 |
0.69 |
| chr3_32993605_32993756 | 0.03 |
CCR4 |
chemokine (C-C motif) receptor 4 |
614 |
0.81 |
| chr18_22811215_22811414 | 0.03 |
ZNF521 |
zinc finger protein 521 |
180 |
0.97 |
| chr17_54374886_54375091 | 0.03 |
ANKFN1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
123452 |
0.06 |
| chr7_50347932_50348149 | 0.03 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
277 |
0.95 |
| chr1_76253803_76253986 | 0.03 |
ENSG00000207241 |
. |
320 |
0.71 |
| chr2_175358586_175358744 | 0.03 |
GPR155 |
G protein-coupled receptor 155 |
6843 |
0.2 |
| chr14_106372328_106372479 | 0.03 |
ENSG00000211925 |
. |
696 |
0.18 |
| chr8_66748245_66748396 | 0.03 |
PDE7A |
phosphodiesterase 7A |
2663 |
0.39 |
| chr22_41080945_41081220 | 0.03 |
MCHR1 |
melanin-concentrating hormone receptor 1 |
5805 |
0.23 |
| chr4_84099158_84099309 | 0.03 |
PLAC8 |
placenta-specific 8 |
41005 |
0.17 |
| chr15_40599321_40599893 | 0.03 |
PLCB2 |
phospholipase C, beta 2 |
419 |
0.68 |
| chr6_31696965_31697269 | 0.02 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
161 |
0.79 |
| chr1_208056644_208056795 | 0.02 |
CD34 |
CD34 molecule |
7118 |
0.29 |
| chr5_79603471_79603622 | 0.02 |
ENSG00000251828 |
. |
1359 |
0.41 |
| chr10_51624082_51624233 | 0.02 |
TIMM23 |
translocase of inner mitochondrial membrane 23 homolog (yeast) |
819 |
0.62 |
| chr3_152001271_152001556 | 0.02 |
MBNL1-AS1 |
MBNL1 antisense RNA 1 |
14069 |
0.21 |
| chr17_62961816_62962168 | 0.02 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
7643 |
0.15 |
| chr4_90031662_90031813 | 0.02 |
TIGD2 |
tigger transposable element derived 2 |
2231 |
0.34 |
| chr6_10528716_10529334 | 0.02 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
436 |
0.83 |
| chr12_116982362_116982513 | 0.02 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
14749 |
0.27 |
| chr20_8113545_8114201 | 0.02 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
571 |
0.84 |
| chr21_16726840_16726991 | 0.02 |
ENSG00000212564 |
. |
259687 |
0.02 |
| chr9_88555446_88555764 | 0.02 |
NAA35 |
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
456 |
0.9 |
| chr12_122327188_122327341 | 0.02 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
572 |
0.69 |
| chr20_47808077_47808228 | 0.02 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
3248 |
0.24 |
| chr18_11851450_11852068 | 0.02 |
CHMP1B |
charged multivesicular body protein 1B |
364 |
0.71 |
| chr19_10346719_10347362 | 0.02 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
5078 |
0.09 |
| chr19_37862447_37862678 | 0.02 |
ZNF527 |
zinc finger protein 527 |
466 |
0.79 |
| chr17_43025339_43025574 | 0.02 |
KIF18B |
kinesin family member 18B |
374 |
0.78 |
| chr7_128002255_128002406 | 0.02 |
PRRT4 |
proline-rich transmembrane protein 4 |
591 |
0.68 |
| chr14_100535763_100535976 | 0.02 |
EVL |
Enah/Vasp-like |
3095 |
0.2 |
| chr7_87506467_87506705 | 0.02 |
SLC25A40 |
solute carrier family 25, member 40 |
922 |
0.44 |
| chr5_130881277_130881428 | 0.02 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
12626 |
0.29 |
| chr2_191746520_191746971 | 0.02 |
AC005540.3 |
|
40 |
0.96 |
| chr15_49102374_49102526 | 0.02 |
CEP152 |
centrosomal protein 152kDa |
341 |
0.57 |
| chr7_8011540_8011691 | 0.02 |
AC006042.7 |
|
2081 |
0.27 |
| chr17_75437754_75438017 | 0.02 |
ENSG00000200651 |
. |
306 |
0.87 |
| chr7_98477525_98477676 | 0.02 |
TRRAP |
transformation/transcription domain-associated protein |
1113 |
0.38 |
| chr19_18770811_18771071 | 0.02 |
KLHL26 |
kelch-like family member 26 |
23074 |
0.1 |
| chr5_72111691_72111929 | 0.02 |
TNPO1 |
transportin 1 |
329 |
0.68 |
| chr2_124669854_124670005 | 0.02 |
ENSG00000223118 |
. |
42782 |
0.2 |
| chr10_111770016_111770167 | 0.02 |
ADD3 |
adducin 3 (gamma) |
2369 |
0.3 |
| chr17_40424757_40425084 | 0.02 |
AC003104.1 |
|
219 |
0.88 |
| chr1_99336669_99336925 | 0.02 |
RP5-896L10.1 |
|
133035 |
0.05 |
| chr7_142013111_142013262 | 0.02 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
23577 |
0.2 |
| chr14_100260916_100261174 | 0.02 |
EML1 |
echinoderm microtubule associated protein like 1 |
1273 |
0.55 |
| chr1_226067206_226067421 | 0.02 |
TMEM63A |
transmembrane protein 63A |
1934 |
0.23 |
| chr11_111895361_111896109 | 0.02 |
DLAT |
dihydrolipoamide S-acetyltransferase |
197 |
0.9 |
| chr11_47572722_47572873 | 0.02 |
CELF1 |
CUGBP, Elav-like family member 1 |
1872 |
0.17 |
| chr19_36395044_36395195 | 0.02 |
HCST |
hematopoietic cell signal transducer |
1679 |
0.17 |
| chr11_2467288_2467521 | 0.02 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
1183 |
0.4 |
| chr19_13319766_13319917 | 0.02 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
48373 |
0.08 |
| chr2_70321192_70321429 | 0.02 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
5332 |
0.17 |
| chrX_47491637_47491788 | 0.02 |
CFP |
complement factor properdin |
2008 |
0.21 |
| chr6_90120799_90121205 | 0.02 |
RRAGD |
Ras-related GTP binding D |
787 |
0.65 |
| chr16_77755925_77756166 | 0.02 |
NUDT7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
366 |
0.87 |
| chr1_235530756_235531012 | 0.02 |
TBCE |
tubulin folding cofactor E |
139 |
0.96 |
| chr17_80858518_80858669 | 0.02 |
TBCD |
tubulin folding cofactor D |
175 |
0.95 |
| chr10_73533824_73534035 | 0.02 |
C10orf54 |
chromosome 10 open reading frame 54 |
674 |
0.7 |
| chr10_75818028_75818179 | 0.02 |
VCL |
vinculin |
25131 |
0.17 |
| chr19_2269030_2269201 | 0.02 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
370 |
0.56 |
| chr7_132766918_132767069 | 0.02 |
CHCHD3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
151 |
0.87 |
| chr1_29255082_29255406 | 0.02 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
14153 |
0.18 |
| chr3_123753408_123753559 | 0.02 |
ENSG00000238512 |
. |
27448 |
0.16 |
| chr16_50315325_50315505 | 0.02 |
ADCY7 |
adenylate cyclase 7 |
1977 |
0.34 |
| chr19_17357282_17357631 | 0.02 |
NR2F6 |
nuclear receptor subfamily 2, group F, member 6 |
707 |
0.42 |
| chr15_69110393_69110544 | 0.02 |
ANP32A |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
2757 |
0.31 |
| chr9_139929883_139930034 | 0.02 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
2496 |
0.09 |
| chr11_62493191_62493431 | 0.02 |
HNRNPUL2 |
heterogeneous nuclear ribonucleoprotein U-like 2 |
1510 |
0.15 |
| chr3_13056686_13056902 | 0.02 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
28258 |
0.23 |
| chr12_63253018_63253169 | 0.02 |
ENSG00000200296 |
. |
8412 |
0.26 |
| chr19_45620700_45620939 | 0.02 |
AC005757.6 |
|
19416 |
0.08 |
| chr17_38947662_38947813 | 0.02 |
KRT28 |
keratin 28 |
8474 |
0.1 |
| chr7_130790301_130790526 | 0.02 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
1376 |
0.46 |
| chr5_39195697_39195848 | 0.02 |
FYB |
FYN binding protein |
7357 |
0.29 |
| chr15_50410621_50410772 | 0.02 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
723 |
0.73 |
| chr19_11457236_11457593 | 0.02 |
CCDC159 |
coiled-coil domain containing 159 |
182 |
0.46 |
| chr1_98385360_98386215 | 0.02 |
DPYD |
dihydropyrimidine dehydrogenase |
766 |
0.78 |
| chr17_2689592_2689743 | 0.02 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
9317 |
0.16 |
| chr1_108464074_108464225 | 0.02 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
42916 |
0.16 |
| chr21_37693064_37693535 | 0.02 |
AP000692.10 |
|
331 |
0.74 |
| chr5_149789702_149789913 | 0.02 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
2488 |
0.26 |
| chr17_66255315_66255711 | 0.02 |
ARSG |
arylsulfatase G |
190 |
0.93 |
| chr11_95418104_95418255 | 0.02 |
FAM76B |
family with sequence similarity 76, member B |
101790 |
0.07 |
| chr9_132000020_132000171 | 0.02 |
ENSG00000220992 |
. |
7099 |
0.18 |
| chr19_44488599_44488754 | 0.02 |
ZNF155 |
zinc finger protein 155 |
303 |
0.82 |
| chr12_122517366_122517604 | 0.02 |
MLXIP |
MLX interacting protein |
857 |
0.66 |
| chr16_85768387_85768538 | 0.02 |
ENSG00000222190 |
. |
6844 |
0.12 |
| chr5_52096731_52097099 | 0.02 |
CTD-2288O8.1 |
|
13055 |
0.19 |
| chr16_48655716_48655879 | 0.02 |
N4BP1 |
NEDD4 binding protein 1 |
11677 |
0.18 |
| chr1_24239672_24239931 | 0.02 |
CNR2 |
cannabinoid receptor 2 (macrophage) |
51 |
0.95 |
| chr22_24537742_24537893 | 0.02 |
CABIN1 |
calcineurin binding protein 1 |
13989 |
0.16 |
| chr5_175963281_175963585 | 0.02 |
RNF44 |
ring finger protein 44 |
988 |
0.42 |
| chr17_7154023_7154644 | 0.02 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
272 |
0.53 |
| chr22_29194035_29194328 | 0.02 |
XBP1 |
X-box binding protein 1 |
1940 |
0.23 |
| chr13_45910861_45911012 | 0.02 |
ENSG00000253051 |
. |
354 |
0.67 |
| chr8_95972695_95972978 | 0.02 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
2600 |
0.21 |
| chr5_102596450_102596781 | 0.02 |
C5orf30 |
chromosome 5 open reading frame 30 |
1490 |
0.46 |
| chr14_24607151_24607302 | 0.02 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
1790 |
0.12 |
| chr9_69262672_69262918 | 0.02 |
CBWD6 |
COBW domain containing 6 |
202 |
0.94 |
| chr2_198173822_198174153 | 0.02 |
ANKRD44 |
ankyrin repeat domain 44 |
1524 |
0.32 |
| chr17_9478666_9479117 | 0.02 |
STX8 |
syntaxin 8 |
263 |
0.83 |
| chr7_6629930_6630111 | 0.02 |
C7orf26 |
chromosome 7 open reading frame 26 |
245 |
0.85 |
| chr10_14643816_14644958 | 0.02 |
FAM107B |
family with sequence similarity 107, member B |
2001 |
0.38 |
| chr12_102018711_102018862 | 0.02 |
RP11-755O11.2 |
|
25940 |
0.14 |
| chr13_78270363_78270592 | 0.02 |
SLAIN1 |
SLAIN motif family, member 1 |
1546 |
0.35 |
| chr5_72144723_72145705 | 0.02 |
TNPO1 |
transportin 1 |
1203 |
0.49 |
| chr3_44802588_44803091 | 0.02 |
KIAA1143 |
KIAA1143 |
315 |
0.52 |
| chr2_64072820_64072971 | 0.02 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
313 |
0.93 |
| chr16_56775385_56775536 | 0.02 |
NUP93 |
nucleoporin 93kDa |
834 |
0.5 |
| chr11_64108374_64108555 | 0.02 |
CCDC88B |
coiled-coil domain containing 88B |
769 |
0.4 |
| chr12_51762995_51763159 | 0.02 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
14270 |
0.14 |
| chr22_37680309_37680627 | 0.02 |
CYTH4 |
cytohesin 4 |
1940 |
0.3 |
| chrX_19362865_19363070 | 0.02 |
PDHA1 |
pyruvate dehydrogenase (lipoamide) alpha 1 |
881 |
0.68 |
| chr20_56008241_56008392 | 0.02 |
RP4-800J21.3 |
|
40198 |
0.12 |
| chr1_229478945_229479176 | 0.02 |
CCSAP |
centriole, cilia and spindle-associated protein |
325 |
0.86 |
| chr9_70856505_70856767 | 0.02 |
CBWD3 |
COBW domain containing 3 |
239 |
0.92 |
| chr9_179160_179373 | 0.02 |
CBWD1 |
COBW domain containing 1 |
194 |
0.94 |
| chr11_84633973_84634163 | 0.02 |
DLG2 |
discs, large homolog 2 (Drosophila) |
156 |
0.97 |
| chr14_93133172_93133323 | 0.01 |
RIN3 |
Ras and Rab interactor 3 |
14401 |
0.25 |
| chr3_107820035_107820186 | 0.01 |
CD47 |
CD47 molecule |
10238 |
0.3 |
| chr11_66138627_66138850 | 0.01 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
553 |
0.52 |
| chr17_38459758_38459909 | 0.01 |
RARA |
retinoic acid receptor, alpha |
5611 |
0.13 |
| chrX_73070691_73070842 | 0.01 |
RP13-216E22.5 |
|
118554 |
0.06 |
| chr12_9144585_9144883 | 0.01 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
68 |
0.97 |
| chr1_227070661_227070872 | 0.01 |
PSEN2 |
presenilin 2 (Alzheimer disease 4) |
1558 |
0.44 |
| chr10_51370591_51371314 | 0.01 |
AGAP8 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
369 |
0.53 |
| chr2_114195012_114195248 | 0.01 |
CBWD2 |
COBW domain containing 2 |
138 |
0.95 |
| chr8_33342029_33342180 | 0.01 |
MAK16 |
MAK16 homolog (S. cerevisiae) |
164 |
0.94 |
| chr9_139737618_139737804 | 0.01 |
C9orf172 |
chromosome 9 open reading frame 172 |
1156 |
0.22 |
| chr19_18429580_18429756 | 0.01 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
2599 |
0.15 |
| chr2_171555186_171555337 | 0.01 |
AC007277.3 |
|
3377 |
0.22 |
| chr7_6673989_6674140 | 0.01 |
ZNF853 |
zinc finger protein 853 |
18816 |
0.11 |
| chr8_9008533_9008971 | 0.01 |
PPP1R3B |
protein phosphatase 1, regulatory subunit 3B |
332 |
0.62 |
| chr20_54989282_54989551 | 0.01 |
CASS4 |
Cas scaffolding protein family member 4 |
2099 |
0.24 |
| chr8_30556332_30556483 | 0.01 |
GSR |
glutathione reductase |
28120 |
0.16 |
| chr13_52397484_52397635 | 0.01 |
RP11-327P2.5 |
|
19126 |
0.17 |
| chrX_102001474_102001892 | 0.01 |
BHLHB9 |
basic helix-loop-helix domain containing, class B, 9 |
661 |
0.72 |
| chr9_70490242_70490494 | 0.01 |
CBWD5 |
COBW domain containing 5 |
122 |
0.98 |
| chr1_1136100_1136251 | 0.01 |
TNFRSF18 |
tumor necrosis factor receptor superfamily, member 18 |
4885 |
0.08 |
| chr10_90750353_90750504 | 0.01 |
FAS |
Fas cell surface death receptor |
14 |
0.92 |
| chr20_46364288_46364439 | 0.01 |
SULF2 |
sulfatase 2 |
49870 |
0.14 |
| chr14_91865794_91865945 | 0.01 |
CCDC88C |
coiled-coil domain containing 88C |
17821 |
0.22 |
| chr6_36880305_36880530 | 0.01 |
C6orf89 |
chromosome 6 open reading frame 89 |
26777 |
0.14 |
| chr14_105885619_105886050 | 0.01 |
RP11-521B24.3 |
|
242 |
0.57 |
| chr14_91866311_91866542 | 0.01 |
CCDC88C |
coiled-coil domain containing 88C |
17264 |
0.22 |
| chr20_62535394_62535545 | 0.01 |
ENSG00000199805 |
. |
4839 |
0.08 |
| chr17_78821534_78821685 | 0.01 |
RP11-28G8.1 |
|
42177 |
0.14 |
| chr7_155131352_155131503 | 0.01 |
BLACE |
B-cell acute lymphoblastic leukemia expressed |
29202 |
0.15 |
| chr6_35014998_35015149 | 0.01 |
TCP11 |
t-complex 11, testis-specific |
73749 |
0.09 |
| chr7_44645646_44645804 | 0.01 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
446 |
0.81 |
| chr5_16465054_16465892 | 0.01 |
ZNF622 |
zinc finger protein 622 |
428 |
0.86 |
| chr11_31391385_31392006 | 0.01 |
DNAJC24 |
DnaJ (Hsp40) homolog, subfamily C, member 24 |
291 |
0.57 |
| chr21_36715747_36715917 | 0.01 |
RUNX1 |
runt-related transcription factor 1 |
294191 |
0.01 |
| chr13_22245209_22245388 | 0.01 |
FGF9 |
fibroblast growth factor 9 |
224 |
0.95 |
| chr19_50003533_50003936 | 0.01 |
hsa-mir-150 |
hsa-mir-150 |
47 |
0.8 |
| chr16_67513595_67513746 | 0.01 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
1312 |
0.23 |
| chr9_36193726_36193938 | 0.01 |
CLTA |
clathrin, light chain A |
2872 |
0.25 |
| chr16_71843274_71843511 | 0.01 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
288 |
0.87 |
| chr4_56812643_56812942 | 0.01 |
CEP135 |
centrosomal protein 135kDa |
2245 |
0.32 |
| chr11_123374923_123375196 | 0.01 |
GRAMD1B |
GRAM domain containing 1B |
21285 |
0.22 |
| chr2_71701457_71701608 | 0.01 |
DYSF |
dysferlin |
7700 |
0.27 |
| chr11_2466894_2467127 | 0.01 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
789 |
0.56 |
| chr15_83419318_83419891 | 0.01 |
RP11-752G15.6 |
|
268 |
0.86 |
| chr1_28196256_28196434 | 0.01 |
THEMIS2 |
thymocyte selection associated family member 2 |
2710 |
0.16 |
| chr1_208041809_208042068 | 0.01 |
CD34 |
CD34 molecule |
21899 |
0.23 |
| chr7_127667736_127667887 | 0.01 |
LRRC4 |
leucine rich repeat containing 4 |
3247 |
0.32 |
| chr11_31831358_31831638 | 0.01 |
PAX6 |
paired box 6 |
355 |
0.87 |
| chr16_70467072_70467318 | 0.01 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
5796 |
0.13 |
| chr22_17567534_17567787 | 0.01 |
IL17RA |
interleukin 17 receptor A |
1811 |
0.31 |
| chr19_42721386_42721863 | 0.01 |
DEDD2 |
death effector domain containing 2 |
197 |
0.87 |
| chr14_22998901_22999052 | 0.01 |
TRAJ15 |
T cell receptor alpha joining 15 |
396 |
0.76 |
| chr17_43664021_43664172 | 0.01 |
RP11-798G7.4 |
|
27660 |
0.11 |
| chr9_92015629_92015819 | 0.01 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
5117 |
0.27 |
| chr16_83761086_83761237 | 0.01 |
RP11-298D21.2 |
|
1179 |
0.45 |
| chr1_154317465_154317703 | 0.01 |
ENSG00000238365 |
. |
6365 |
0.11 |
| chr1_172106062_172106431 | 0.01 |
ENSG00000207949 |
. |
1801 |
0.34 |
| chr1_32715929_32716174 | 0.01 |
LCK |
lymphocyte-specific protein tyrosine kinase |
789 |
0.4 |
| chr8_19553922_19554073 | 0.01 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
13725 |
0.29 |
| chr9_115248402_115248939 | 0.01 |
KIAA1958 |
KIAA1958 |
457 |
0.66 |
| chr3_93781843_93782253 | 0.01 |
DHFRL1 |
dihydrofolate reductase-like 1 |
19 |
0.8 |
| chr5_130598985_130599244 | 0.01 |
CDC42SE2 |
CDC42 small effector 2 |
588 |
0.84 |
| chr7_158496384_158496597 | 0.01 |
NCAPG2 |
non-SMC condensin II complex, subunit G2 |
967 |
0.62 |
| chr17_61629421_61629699 | 0.01 |
DCAF7 |
DDB1 and CUL4 associated factor 7 |
1699 |
0.27 |
| chr11_71791059_71791281 | 0.01 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
212 |
0.52 |
| chr14_22959148_22959353 | 0.01 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
3079 |
0.14 |
| chr11_11642887_11643184 | 0.01 |
GALNT18 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
517 |
0.82 |
| chr9_127976807_127976958 | 0.01 |
RABEPK |
Rab9 effector protein with kelch motifs |
13998 |
0.14 |
| chr18_48405507_48405704 | 0.01 |
ME2 |
malic enzyme 2, NAD(+)-dependent, mitochondrial |
115 |
0.97 |
| chr16_27241764_27241915 | 0.01 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
2560 |
0.25 |
| chr2_121268328_121268479 | 0.01 |
LINC01101 |
long intergenic non-protein coding RNA 1101 |
44706 |
0.19 |
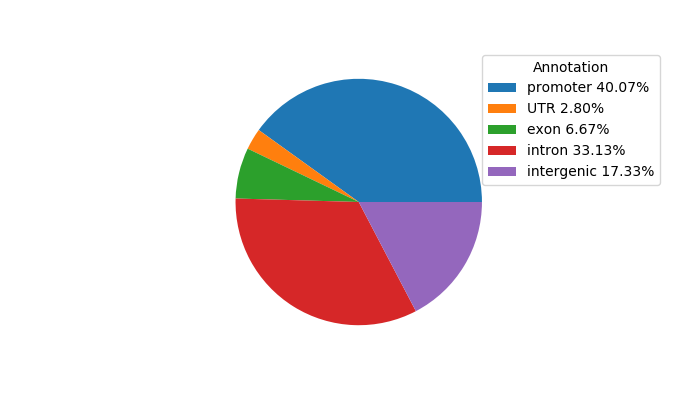
Network of associatons between targets according to the STRING database.